Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354
- PMID: 20956322
- PMCID: PMC2973864
- DOI: 10.1073/pnas.1011036107
Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354
Abstract
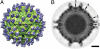
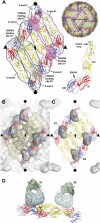
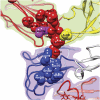
Many flaviviruses are significant human pathogens, with the humoral immune response playing an essential role in restricting infection and disease. CR4354, a human monoclonal antibody isolated from a patient, neutralizes West Nile virus (WNV) infection at a postattachment stage in the viral life-cycle. Here, we determined the structure of WNV complexed with Fab fragments of CR4354 using cryoelectron microscopy. The outer glycoprotein shell of a mature WNV particle is formed by 30 rafts of three homodimers of the viral surface protein E. CR4354 binds to a discontinuous epitope formed by protein segments from two neighboring E molecules, but does not cause any detectable structural disturbance on the viral surface. The epitope occurs at two independent positions within an icosahedral asymmetric unit, resulting in 120 binding sites on the viral surface. The cross-linking of the six E monomers within one raft by four CR4354 Fab fragments suggests that the antibody neutralizes WNV by blocking the pH-induced rearrangement of the E protein required for virus fusion with the endosomal membrane.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

