B-cell activating factor and v-Myc myelocytomatosis viral oncogene homolog (c-Myc) influence progression of chronic lymphocytic leukemia
- PMID: 20956327
- PMCID: PMC2973856
- DOI: 10.1073/pnas.1013420107
B-cell activating factor and v-Myc myelocytomatosis viral oncogene homolog (c-Myc) influence progression of chronic lymphocytic leukemia
Abstract
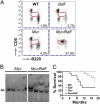
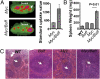
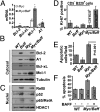
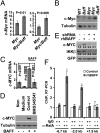
Mice bearing a v-Myc myelocytomatosis viral oncogene homolog (c-Myc) transgene controlled by an Ig-alpha heavy-chain enhancer (iMyc(Cα) mice) rarely develop lymphomas but instead have increased rates of memory B-cell turnover and impaired antibody responses to antigen. We found that male progeny of iMyc(Cα) mice mated with mice transgenic (Tg) for CD257 (B-cell activating factor, BAFF) developed CD5(+) B-cell leukemia resembling human chronic lymphocytic leukemia (CLL), which also displays a male gender bias. Surprisingly, leukemic cells of Myc/Baff Tg mice expressed higher levels of c-Myc than did B cells of iMyc(Cα) mice. We found that CLL cells of many patients with progressive disease also expressed high amounts of c-MYC, particularly CLL cells whose survival depends on nurse-like cells (NLC), which express high-levels of BAFF. We find that BAFF could enhance CLL-cell expression of c-MYC via activation the canonical IκB kinase (IKK)/NF-κB pathway. Inhibition of the IKK/NF-κB pathway in mouse or human leukemia cells blocked the capacity of BAFF to induce c-MYC or promote leukemia-cell survival and significantly impaired disease progression in Myc/Baff Tg mice. This study reveals an important relationship between BAFF and c-MYC in CLL which may affect disease development and progression, and suggests that inhibitors of the canonical NF-κB pathway may be effective in treatment of patients with this disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Chronic lymphocytic leukemia of Emu-TCL1 transgenic mice undergoes rapid cell turnover that can be offset by extrinsic CD257 to accelerate disease progression.Blood. 2009 Nov 12;114(20):4469-76. doi: 10.1182/blood-2009-06-230169. Epub 2009 Sep 15. Blood. 2009. PMID: 19755673
-
Critical role of B cell lymphoma 10 in BAFF-regulated NF-κB activation and survival of anergic B cells.J Immunol. 2012 Dec 1;189(11):5185-93. doi: 10.4049/jimmunol.1102952. Epub 2012 Oct 19. J Immunol. 2012. PMID: 23087406 Free PMC article.
-
BAFF and APRIL support chronic lymphocytic leukemia B-cell survival through activation of the canonical NF-kappaB pathway.Blood. 2007 Jan 15;109(2):703-10. doi: 10.1182/blood-2006-06-027755. Epub 2006 Sep 14. Blood. 2007. PMID: 16973958 Free PMC article.
-
BAFF signaling in health and disease.Curr Opin Immunol. 2021 Aug;71:124-131. doi: 10.1016/j.coi.2021.06.014. Epub 2021 Aug 2. Curr Opin Immunol. 2021. PMID: 34352467 Review.
-
The multi-faceted roles of MYC in the prognosis of chronic lymphocytic leukemia.Leuk Lymphoma. 2025 May;66(5):805-817. doi: 10.1080/10428194.2024.2447362. Epub 2025 Jan 2. Leuk Lymphoma. 2025. PMID: 39743868 Review.
Cited by
-
Paracrine WNT5A Signaling Inhibits Expansion of Tumor-Initiating Cells.Cancer Res. 2015 May 15;75(10):1972-82. doi: 10.1158/0008-5472.CAN-14-2761. Epub 2015 Mar 13. Cancer Res. 2015. PMID: 25769722 Free PMC article.
-
Inherited susceptibility to chronic lymphocytic leukemia: evidence and prospects for the future.Ther Adv Hematol. 2013 Aug;4(4):298-308. doi: 10.1177/2040620713495639. Ther Adv Hematol. 2013. PMID: 23926461 Free PMC article.
-
Biology of chronic lymphocytic leukemia in different microenvironments: clinical and therapeutic implications.Hematol Oncol Clin North Am. 2013 Apr;27(2):173-206. doi: 10.1016/j.hoc.2013.01.002. Hematol Oncol Clin North Am. 2013. PMID: 23561469 Free PMC article. Review.
-
Increased aldehyde dehydrogenase activity in high-risk chronic lymphocytic leukemia.Leuk Lymphoma. 2013 Feb;54(2):400-2. doi: 10.3109/10428194.2012.709630. Epub 2012 Aug 13. Leuk Lymphoma. 2013. PMID: 22784365 Free PMC article. No abstract available.
-
CX-3543 Promotes Cell Apoptosis through Downregulation of CCAT1 in Colon Cancer Cells.Biomed Res Int. 2018 Nov 4;2018:9701957. doi: 10.1155/2018/9701957. eCollection 2018. Biomed Res Int. 2018. PMID: 30519593 Free PMC article.
References
-
- Ladanyi M, Offit K, Jhanwar SC, Filippa DA, Chaganti RS. MYC rearrangement and translocations involving band 8q24 in diffuse large cell lymphomas. Blood. 1991;77:1057–1063. - PubMed
-
- Avet-Loiseau H, et al. Intergroupe Francophone du Myélome. Rearrangements of the c-myc oncogene are present in 15% of primary human multiple myeloma tumors. Blood. 2001;98:3082–3086. - PubMed
-
- Huh YO, et al. MYC translocation in chronic lymphocytic leukaemia is associated with increased prolymphocytes and a poor prognosis. Br J Haematol. 2008;142:36–44. - PubMed
-
- Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

