Major differences in the responses of primary human leukocyte subsets to IFN-beta
- PMID: 20956346
- PMCID: PMC3244975
- DOI: 10.4049/jimmunol.0902314
Major differences in the responses of primary human leukocyte subsets to IFN-beta
Abstract
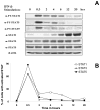
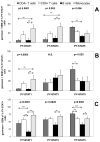
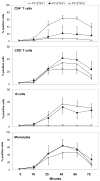
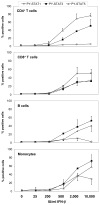
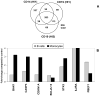
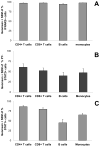
Treatment of cell lines with type I IFNs activates the formation of IFN-stimulated gene factor 3 (STAT1/STAT2/IFN regulatory factor-9), which induces the expression of many genes. To study this response in primary cells, we treated fresh human blood with IFN-β and used flow cytometry to analyze phosphorylated STAT1, STAT3, and STAT5 in CD4(+) and CD8(+) T cells, B cells, and monocytes. The activation of STAT1 was remarkably different among these leukocyte subsets. In contrast to monocytes and CD4(+) and CD8(+) T cells, few B cells activated STAT1 in response to IFN-β, a finding that could not be explained by decreased levels of IFNAR2 or STAT1 or enhanced levels of suppressor of cytokine signaling 1 or relevant protein tyrosine phosphatases in B cells. Microarray and real-time PCR analyses revealed the induction of STAT1-dependent proapoptotic mRNAs in monocytes but not in B cells. These data show that IFN-stimulated gene factor 3 or STAT1 homodimers are not the main activators of gene expression in primary B cells of healthy humans. Notably, in B cells and, especially in CD4(+) T cells, IFN-β activated STAT5 in addition to STAT3, with biological effects often opposite from those driven by activated STAT1. These data help to explain why IFN-β increases the survival of primary human B cells and CD4(+) T cells but enhances the apoptosis of monocytes, as well as to understand how leukocyte subsets are differentially affected by endogenous type I IFNs during viral or bacterial infections and by type I IFN treatment of patients with multiple sclerosis, hepatitis, or cancer.
Figures
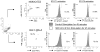


Similar articles
-
The role of cell type-specific responses in IFN-β therapy of multiple sclerosis.Proc Natl Acad Sci U S A. 2011 Dec 6;108(49):19689-94. doi: 10.1073/pnas.1117347108. Epub 2011 Nov 21. Proc Natl Acad Sci U S A. 2011. PMID: 22106296 Free PMC article.
-
Three Copies of Four Interferon Receptor Genes Underlie a Mild Type I Interferonopathy in Down Syndrome.J Clin Immunol. 2020 Aug;40(6):807-819. doi: 10.1007/s10875-020-00803-9. Epub 2020 Jun 22. J Clin Immunol. 2020. PMID: 32572726 Free PMC article.
-
Elevated STAT1 expression but not phosphorylation in lupus B cells correlates with disease activity and increased plasmablast susceptibility.Rheumatology (Oxford). 2020 Nov 1;59(11):3435-3442. doi: 10.1093/rheumatology/keaa187. Rheumatology (Oxford). 2020. PMID: 32357246
-
A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses.Front Immunol. 2018 May 28;9:1135. doi: 10.3389/fimmu.2018.01135. eCollection 2018. Front Immunol. 2018. PMID: 29892288 Free PMC article. Review.
-
The role of STAT5 in lymphocyte development and transformation.Curr Opin Immunol. 2012 Apr;24(2):146-52. doi: 10.1016/j.coi.2012.01.015. Epub 2012 Feb 16. Curr Opin Immunol. 2012. PMID: 22342169 Free PMC article. Review.
Cited by
-
Elevated type I interferon-like activity in a subset of multiple sclerosis patients: molecular basis and clinical relevance.J Neuroinflammation. 2012 Jun 22;9:140. doi: 10.1186/1742-2094-9-140. J Neuroinflammation. 2012. PMID: 22727118 Free PMC article.
-
Recent Advances of Type I Interferon on the Regulation of Immune Cells and the Treatment of Systemic Lupus Erythematosus.J Inflamm Res. 2025 Mar 30;18:4533-4549. doi: 10.2147/JIR.S516195. eCollection 2025. J Inflamm Res. 2025. PMID: 40182060 Free PMC article. Review.
-
Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation.Immunol Rev. 2012 Nov;250(1):317-34. doi: 10.1111/imr.12001. Immunol Rev. 2012. PMID: 23046138 Free PMC article. Review.
-
Multiplexed Gene Expression as a Characterization of Bioactivity for Interferon Beta (IFN-β) Biosimilar Candidates: Impact of Innate Immune Response Modulating Impurities (IIRMIs).AAPS J. 2019 Feb 8;21(2):26. doi: 10.1208/s12248-019-0300-7. AAPS J. 2019. PMID: 30737590
-
The molecular basis for differential type I interferon signaling.J Biol Chem. 2017 May 5;292(18):7285-7294. doi: 10.1074/jbc.R116.774562. Epub 2017 Mar 13. J Biol Chem. 2017. PMID: 28289098 Free PMC article. Review.
References
-
- Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. - PubMed
-
- Novick D, Cohen B, Rubinstein M. The human interferon alpha/beta receptor: characterization and molecular cloning. Cell. 1994;77:391–400. - PubMed
-
- Stark GR, I, Kerr M, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. - PubMed
-
- van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. - PubMed
-
- Decker T, Kovarik P, Meinke A. GAS elements: a few nucleotides witha major impact on cytokine-induced gene expression. J Interferon Cytokine Res. 1997;17:121–134. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

