Tracking origins and spread of sulfadoxine-resistant Plasmodium falciparum dhps alleles in Thailand
- PMID: 20956597
- PMCID: PMC3019650
- DOI: 10.1128/AAC.00691-10
Tracking origins and spread of sulfadoxine-resistant Plasmodium falciparum dhps alleles in Thailand
Abstract
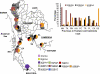
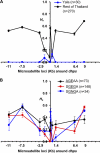
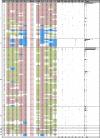
The emergence and spread of drug-resistant Plasmodium falciparum have been a major impediment for the control of malaria worldwide. Earlier studies have shown that similar to chloroquine (CQ) resistance, high levels of pyrimethamine resistance in P. falciparum originated independently 4 to 5 times globally, including one origin at the Thailand-Cambodia border. In this study we describe the origins and spread of sulfadoxine-resistance-conferring dihydropteroate synthase (dhps) alleles in Thailand. The dhps mutations and flanking microsatellite loci were genotyped for P. falciparum isolates collected from 11 Thai provinces along the Burma, Cambodia, and Malaysia borders. Results indicated that resistant dhps alleles were fixed in Thailand, predominantly being the SGEGA, AGEAA, and SGNGA triple mutants and the AGKAA double mutant (mutated codons are underlined). These alleles had different geographical distributions. The SGEGA alleles were found mostly at the Burma border, while the SGNGA alleles occurred mainly at the Cambodia border and nearby provinces. Microsatellite data suggested that there were two major genetic lineages of the triple mutants in Thailand, one common for SGEGA/SGNGA alleles and another one independent for AGEAA. Importantly, the newly reported SGNGA alleles possibly originated at the Thailand-Cambodia border. All parasites in the Yala province (Malaysia border) had AGKAA alleles with almost identical flanking microsatellites haplotypes. They were also identical at putatively neutral loci on chromosomes 2 and 3, suggesting a clonal nature of the parasite population in Yala. In summary, this study suggests multiple and independent origins of resistant dhps alleles in Thailand.
Figures

References
-
- Anderson, T. J., and C. Roper. 2005. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop. 94:269-280. - PubMed
-
- Babiker, H. A., I. M. Hastings, and G. Swedberg. 2009. Impaired fitness of drug-resistant malaria parasites: evidence and implication on drug-deployment policies. Expert Rev. Anti Infect. Ther. 7:581-593. - PubMed
-
- Biswas, S., A. Escalante, S. Chaiyaroj, P. Angkasekwinai, and A. A. Lal. 2000. Prevalence of point mutations in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum isolates from India and Thailand: a molecular epidemiologic study. Trop. Med. Int. Health 5:737-743. - PubMed
-
- Brooks, D. R., P. Wang, M. Read, W. M. Watkins, P. F. Sims, and J. E. Hyde. 1994. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase:dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur. J. Biochem. 224:397-405. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

