Use of ITS2 region as the universal DNA barcode for plants and animals
- PMID: 20957043
- PMCID: PMC2948509
- DOI: 10.1371/journal.pone.0013102
Use of ITS2 region as the universal DNA barcode for plants and animals
Abstract
Background: The internal transcribed spacer 2 (ITS2) region of nuclear ribosomal DNA is regarded as one of the candidate DNA barcodes because it possesses a number of valuable characteristics, such as the availability of conserved regions for designing universal primers, the ease of its amplification, and sufficient variability to distinguish even closely related species. However, a general analysis of its ability to discriminate species in a comprehensive sample set is lacking.
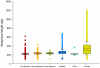
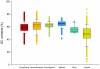
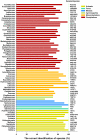
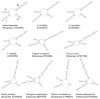
Methodology/principal findings: In the current study, 50,790 plant and 12,221 animal ITS2 sequences downloaded from GenBank were evaluated according to sequence length, GC content, intra- and inter-specific divergence, and efficiency of identification. The results show that the inter-specific divergence of congeneric species in plants and animals was greater than its corresponding intra-specific variations. The success rates for using the ITS2 region to identify dicotyledons, monocotyledons, gymnosperms, ferns, mosses, and animals were 76.1%, 74.2%, 67.1%, 88.1%, 77.4%, and 91.7% at the species level, respectively. The ITS2 region unveiled a different ability to identify closely related species within different families and genera. The secondary structure of the ITS2 region could provide useful information for species identification and could be considered as a molecular morphological characteristic.
Conclusions/significance: As one of the most popular phylogenetic markers for eukaryota, we propose that the ITS2 locus should be used as a universal DNA barcode for identifying plant species and as a complementary locus for CO1 to identify animal species. We have also developed a web application to facilitate ITS2-based cross-kingdom species identification (http://its2-plantidit.dnsalias.org).
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

