µFBI: a microfluidic bead-based immunoassay for multiplexed detection of proteins from a µL sample volume
- PMID: 20957050
- PMCID: PMC2948516
- DOI: 10.1371/journal.pone.0013125
µFBI: a microfluidic bead-based immunoassay for multiplexed detection of proteins from a µL sample volume
Abstract
Background: Over the last ten years, miniaturized multiplexed immunoassays have become robust, reliable research tools that enable researchers to simultaneously determine a multitude of parameters. Among the numerous analytical protein arrays available, bead-based assay systems have evolved into a key technology that enables the quantitative protein profiling of biological samples whilst requiring only a minimal amount of sample material.
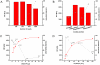
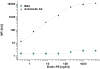
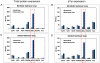
Methodology/principal findings: A microfluidic bead-based immunoassay, µFBI, was developed to perform bead-based multiplexed sandwich immunoassays in a capillary. This setup allows the simultaneous detection of several parameters and only requires 200 ng of tissue lysate in a 1 µL assay volume. In addition, only 1 µL of detection antibodies and 1 µL of the reporter molecule Streptavidin-Phycoerythrin were required. The µFBI was used to compare the expression of seven receptor tyrosine kinases and their degree of tyrosine phosphorylation in breast cancer tissue and in normal tissue lysates. The total amount of HER-2, as well the degree of tyrosine phosphorylation was much higher in breast cancer tissue than in normal tissue. µFBI and a standard bead-based assay led to identical protein expression data. Moreover, it was possible to reduce the quantity of sample material required by a factor of 100 and the quantity of reagents by a factor of 30.
Conclusions/significance: The µFBI, microfluidic bead-based immunoassay, allows the analysis of multiple parameters from a very small amount of sample material, such as tumor biopsies or tissue sections.
Conflict of interest statement
Figures


Similar articles
-
An integrated microfluidic biochemical detection system for protein analysis with magnetic bead-based sampling capabilities.Lab Chip. 2002 Feb;2(1):27-30. doi: 10.1039/b107540n. Epub 2001 Dec 6. Lab Chip. 2002. PMID: 15100857
-
Multiplexed immunoassays for the analysis of breast cancer biopsies.Anal Bioanal Chem. 2010 Aug;397(8):3329-38. doi: 10.1007/s00216-010-3873-7. Epub 2010 Jul 18. Anal Bioanal Chem. 2010. PMID: 20640897
-
Transposing Lateral Flow Immunoassays to Capillary-Driven Microfluidics Using Self-Coalescence Modules and Capillary-Assembled Receptor Carriers.Anal Chem. 2020 Jan 7;92(1):940-946. doi: 10.1021/acs.analchem.9b03792. Epub 2019 Dec 20. Anal Chem. 2020. PMID: 31860276
-
Immunoassays in microfluidic systems.Anal Bioanal Chem. 2010 Jun;397(3):991-1007. doi: 10.1007/s00216-010-3678-8. Epub 2010 Apr 27. Anal Bioanal Chem. 2010. PMID: 20422163 Review.
-
Microfluidic-based blood immunoassays.J Pharm Biomed Anal. 2023 May 10;228:115313. doi: 10.1016/j.jpba.2023.115313. Epub 2023 Feb 24. J Pharm Biomed Anal. 2023. PMID: 36868029 Review.
Cited by
-
Micromotor-Based Biosensing Using Directed Transport of Functionalized Beads.ACS Sens. 2020 Apr 24;5(4):936-942. doi: 10.1021/acssensors.9b02041. Epub 2020 Mar 24. ACS Sens. 2020. PMID: 32141739 Free PMC article.
-
Fiber composite slices for multiplexed immunoassays.Biomicrofluidics. 2015 Jul 29;9(4):044109. doi: 10.1063/1.4927590. eCollection 2015 Jul. Biomicrofluidics. 2015. PMID: 26339310 Free PMC article.
-
Protein Microarrays with Novel Microfluidic Methods: Current Advances.Microarrays (Basel). 2014 Jul 1;3(3):180-202. doi: 10.3390/microarrays3030180. Microarrays (Basel). 2014. PMID: 27600343 Free PMC article. Review.
-
Surface Functionalization Methods to Enhance Bioconjugation in Metal-Labeled Polystyrene Particles.Macromolecules. 2011 Jun 28;44(12):4801-4813. doi: 10.1021/ma200582q. Macromolecules. 2011. PMID: 21799543 Free PMC article.
-
Development and validation of a microfluidic immunoassay capable of multiplexing parallel samples in microliter volumes.Lab Chip. 2015 Aug 7;15(15):3211-21. doi: 10.1039/c5lc00398a. Lab Chip. 2015. PMID: 26130452 Free PMC article.
References
-
- Kricka LJ, Master SR, Joos TO, Fortina P. Current perspectives in protein array technology. Ann Clin Biochem. 2006;43:457–467. - PubMed
-
- Wulfkuhle JD, Speer R, Pierobon M, Laird J, Espina V, et al. Multiplexed cell signaling analysis of human breast cancer applications for personalized therapy. J Proteome Res. 2008;7:1508–1517. - PubMed
-
- Qiu J, Hanash S. Autoantibody profiling for cancer detection. Clin Lab Med. 2009;29:31–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

