Chitosanase-based method for RNA isolation from cells transfected with chitosan/siRNA nanocomplexes for real-time RT-PCR in gene silencing
- PMID: 20957169
- PMCID: PMC2950405
- DOI: 10.2147/ijn.s10879
Chitosanase-based method for RNA isolation from cells transfected with chitosan/siRNA nanocomplexes for real-time RT-PCR in gene silencing
Abstract
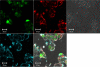
Chitosan, a well known natural cationic polysaccharide, has been successfully implemented in vitro and in vivo as a nonviral delivery system for both plasmid DNA and siRNA. While using chitosan/siRNA polyplexes to knock down specific targets, we have underestimated the effect of nucleic acids binding to chitosan when extracting RNA for subsequent quantitative PCR evaluation of silencing. In vitro transfection using chitosan/siRNA-based polyplexes reveals a very poor recovery of total RNA especially when using low cell numbers in 96 well plates. Here, we describe a method that dramatically enhances RNA extraction from chitosan/siRNA-treated cells by using an enzymatic treatment with a type III chitosanase. We show that chitosanase treatment prior to RNA extraction greatly enhances the yield and the integrity of extracted RNA. This method will therefore eliminate the bias associated with lower RNA yield and integrity when quantifying gene silencing of chitosan-based systems using quantitative real time PCR.
Keywords: DPP-IV gene silencing; RIN; chitosan; chitosanase; qPCR; siRNA.
Figures
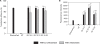



References
-
- Illum L. Chitosan and its use as a pharmaceutical excipient. Pharm Res. 1998;15:1326–1331. - PubMed
-
- Huang M, Khor E, Lim LY. Uptake and cytotoxicity of chitosan molecules and nanoparticles: effects of molecular weight and degree of deacetylation. Pharm Res. 2004;21:344–353. - PubMed
-
- Zhang H, Neau SH. In vitro degradation of chitosan by a commercial enzyme preparation: effect of molecular weight and degree of deacetylation. Biomaterials. 2001;22:1653–1658. - PubMed
-
- Prego C, Torres D, Alonso MJ. Chitosan nanocapsules as carriers for oral peptide delivery: effect of chitosan molecular weight and type of salt on the in vitro behaviour and in vivo effectiveness. J Nanosci Nanotechnol. 2006;6:2921–2928. - PubMed
-
- Prego C, Fabre M, Torres D, Alonso MJ. Efficacy and mechanism of action of chitosan nanocapsules for oral peptide delivery. Pharm Res. 2006;23:549–556. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

