A network of cancer genes with co-occurring and anti-co-occurring mutations
- PMID: 20957180
- PMCID: PMC2949398
- DOI: 10.1371/journal.pone.0013180
A network of cancer genes with co-occurring and anti-co-occurring mutations
Abstract
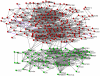
Certain cancer genes contribute to tumorigenesis in a manner of either co-occurring or mutually exclusive (anti-co-occurring) mutations; however, the global picture of when, where and how these functional interactions occur remains unclear. This study presents a systems biology approach for this purpose. After applying this method to cancer gene mutation data generated from large-scale and whole genome sequencing of cancer samples, a network of cancer genes with co-occurring and anti-co-occurring mutations was constructed. Analysis of this network revealed that genes with co-occurring mutations prefer direct signaling transductions and that the interaction relations among cancer genes in the network are related with their functional similarity. It was also revealed that genes with co-occurring mutations tend to have similar mutation frequencies, whereas genes with anti-co-occurring mutations tend to have different mutation frequencies. Moreover, genes with more exons tend to have more co-occurring mutations with other genes, and genes having lower local coherent network structures tend to have higher mutation frequency. The network showed two complementary modules that have distinct functions and have different roles in tumorigenesis. This study presented a framework for the analysis of cancer genome sequencing outputs. The presented data and uncovered patterns are helpful for understanding the contribution of gene mutations to tumorigenesis and valuable in the identification of key biomarkers and drug targets for cancer.
Conflict of interest statement
Figures


References
-
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

