The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase
- PMID: 20957187
- PMCID: PMC2950127
- DOI: 10.1371/journal.pbio.1000496
The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase
Abstract
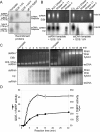
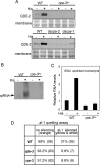
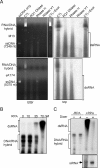
The production of aberrant RNA (aRNA) is the initial step in several RNAi pathways. How aRNA is produced and specifically recognized by RNA-dependent RNA polymerases (RdRPs) to generate double-stranded RNA (dsRNA) is not clear. We previously showed that in the filamentous fungus Neurospora, the RdRP QDE-1 is required for rDNA-specific aRNA production, suggesting that QDE-1 may be important in aRNA synthesis. Here we show that a recombinant QDE-1 is both an RdRP and a DNA-dependent RNA polymerase (DdRP). Its DdRP activity is much more robust than the RdRP activity and occurs on ssDNA but not dsDNA templates. We further show that Replication Protein A (RPA), a single-stranded DNA-binding complex that interacts with QDE-1, is essential for aRNA production and gene silencing. In vitro reconstitution assays demonstrate that QDE-1 can produce dsRNA from ssDNA, a process that is strongly promoted by RPA. Furthermore, the interaction between QDE-1 and RPA requires the RecQ DNA helicase QDE-3, a homolog of the human Werner/Bloom Syndrome proteins. Together, these results suggest a novel small RNA biogenesis pathway in Neurospora and a new mechanism for the production of aRNA and dsRNA in RNAi pathways.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14:1041–1048. - PubMed
-
- Catalanotto C, Nolan T, Cogoni C. Homology effects in Neurospora crassa. FEMS Microbiol Lett. 2006;254:182–189. - PubMed
-
- Hannon G. J. RNA interference. Nature. 2002;418:244–251. - PubMed
-
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. - PubMed
-
- Mello C. C, Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

