The constitutive activation of Jak2-V617F is mediated by a π stacking mechanism involving phenylalanines 595 and 617
- PMID: 20958061
- PMCID: PMC2982877
- DOI: 10.1021/bi1014858
The constitutive activation of Jak2-V617F is mediated by a π stacking mechanism involving phenylalanines 595 and 617
Abstract
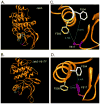
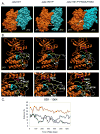
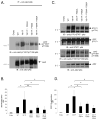
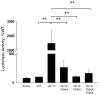
Somatic mutations in the Jak2 allele that lead to constitutive kinase activation of the protein have been identified in human disease conditions such as the myeloproliferative neoplasms (MPNs). The most common mutation in these patients is a V617F substitution mutation, which is believed to play a causative role in the MPN pathogenesis. As such, identifying the molecular basis for the constitutive activation of Jak2-V617F is important for understanding its clinical implications and potential treatment. Here, we hypothesized that conversion of residue 617 from Val to Phe resulted in the formation of novel π stacking interactions with neighboring Phe residues. To test this, we first examined the Jak2 structure via molecular modeling and identified a potential π stacking interaction between F594, F595, and F617. Disruption of this interaction through site-directed mutagenesis impaired Jak2 autophosphorylation, Jak2-dependent gene transcription, and in vitro kinase activity of the Jak2-V617F protein. Further, substitution of F594 and F595 with Trp did not affect Jak2 function significantly, but replacement with charged residues did, showing the importance of aromaticity and hydropathy index conservation at these positions. Using molecular dynamics (MD) simulations, we found that the π stacking interaction between residues 595 and 617 in the Jak2-V617F protein was of much greater energy and conformed to the properties of π stacking, relative to the Jak2-WT or Jak2-V617F/F594A/F595A. In summary, we have identified a π stacking interaction between F595 and F617 that is specific to and is critical for the constitutive activation of Jak2-V617F.
Figures


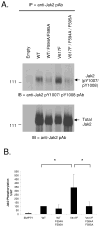
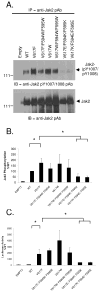
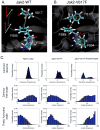
Similar articles
-
A shift in the salt bridge interaction of residues D620 and E621 mediates the constitutive activation of Jak2-H538Q/K539L.Mol Cell Biochem. 2012 Aug;367(1-2):125-40. doi: 10.1007/s11010-012-1326-7. Epub 2012 May 15. Mol Cell Biochem. 2012. PMID: 22584586
-
JAK2 V617F constitutive activation requires JH2 residue F595: a pseudokinase domain target for specific inhibitors.PLoS One. 2010 Jun 16;5(6):e11157. doi: 10.1371/journal.pone.0011157. PLoS One. 2010. PMID: 20585391 Free PMC article.
-
Mechanisms of constitutive activation of Janus kinase 2-V617F revealed at the atomic level through molecular dynamics simulations.Cancer. 2009 Apr 15;115(8):1692-700. doi: 10.1002/cncr.24183. Cancer. 2009. PMID: 19195039
-
JAK2 V617F and beyond: role of genetics and aberrant signaling in the pathogenesis of myeloproliferative neoplasms.Expert Rev Hematol. 2010 Jun;3(3):323-37. doi: 10.1586/ehm.10.28. Expert Rev Hematol. 2010. PMID: 21082983 Review.
-
[Analysis of oncogenic signaling pathway induced by a myeloproliferative neoplasm-associated Janus kinase 2 (JAK2) V617F mutant].Yakugaku Zasshi. 2012;132(11):1267-72. doi: 10.1248/yakushi.12-00225. Yakugaku Zasshi. 2012. PMID: 23123718 Review. Japanese.
Cited by
-
Characterization of JAK1 Pseudokinase Domain in Cytokine Signaling.Cancers (Basel). 2019 Dec 27;12(1):78. doi: 10.3390/cancers12010078. Cancers (Basel). 2019. PMID: 31892268 Free PMC article.
-
Structure of a pseudokinase-domain switch that controls oncogenic activation of Jak kinases.Nat Struct Mol Biol. 2013 Oct;20(10):1221-3. doi: 10.1038/nsmb.2673. Epub 2013 Sep 8. Nat Struct Mol Biol. 2013. PMID: 24013208 Free PMC article.
-
Janus kinase inhibitors: jackpot or potluck?Oncol Rev. 2012 Jun 20;6(1):e13. doi: 10.4081/oncol.2012.e13. eCollection 2012 Mar 5. Oncol Rev. 2012. PMID: 25992203 Free PMC article. Review.
-
ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation.Proc Natl Acad Sci U S A. 2015 Apr 14;112(15):4642-7. doi: 10.1073/pnas.1423201112. Epub 2015 Mar 30. Proc Natl Acad Sci U S A. 2015. PMID: 25825724 Free PMC article.
-
Janus kinase 2 activation mechanisms revealed by analysis of suppressing mutations.J Allergy Clin Immunol. 2019 Apr;143(4):1549-1559.e6. doi: 10.1016/j.jaci.2018.07.022. Epub 2018 Aug 6. J Allergy Clin Immunol. 2019. PMID: 30092288 Free PMC article.
References
-
- Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. - PubMed
-
- Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G, Ihle JN. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. - PubMed
-
- Sandberg EM, Wallace TA, Godeny MD, VonDerLinden D, Sayeski PP. Jak2 tyrosine kinase: a true jak of all trades? Cell Biochem Biophys. 2004;41:207–232. - PubMed
-
- Witthuhn BA, Quelle FW, Silvennoinen O, Yi T, Tang B, Miura O, Ihle JN. JAK2 associates with the erythropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell. 1993;74:227–236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

