The imprint of the Slave Trade in an African American population: mitochondrial DNA, Y chromosome and HTLV-1 analysis in the Noir Marron of French Guiana
- PMID: 20958967
- PMCID: PMC2973943
- DOI: 10.1186/1471-2148-10-314
The imprint of the Slave Trade in an African American population: mitochondrial DNA, Y chromosome and HTLV-1 analysis in the Noir Marron of French Guiana
Abstract
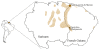
Background: Retracing the genetic histories of the descendant populations of the Slave Trade (16th-19th centuries) is particularly challenging due to the diversity of African ethnic groups involved and the different hybridisation processes with Europeans and Amerindians, which have blurred their original genetic inheritances. The Noir Marron in French Guiana are the direct descendants of maroons who escaped from Dutch plantations in the current day Surinam. They represent an original ethnic group with a highly blended culture. Uniparental markers (mtDNA and NRY) coupled with HTLV-1 sequences (env and LTR) were studied to establish the genetic relationships linking them to African American and African populations.
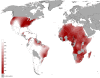
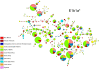
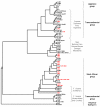
Results: All genetic systems presented a high conservation of the African gene pool (African ancestry: mtDNA = 99.3%; NRY = 97.6%; HTLV-1 env = 20/23; HTLV-1 LTR = 6/8). Neither founder effect nor genetic drift was detected and the genetic diversity is within a range commonly observed in Africa. Higher genetic similarities were observed with the populations inhabiting the Bight of Benin (from Ivory Coast to Benin). Other ancestries were identified but they presented an interesting sex-bias. Whilst male origins spread throughout the north of the bight (from Benin to Senegal), female origins were spread throughout the south (from the Ivory Coast to Angola).
Conclusions: The Noir Marron are unique in having conserved their African genetic ancestry, despite major cultural exchanges with Amerindians and Europeans through inhabiting the same region for four centuries. Their maroon identity and the important number of slaves deported in this region have maintained the original African diversity. All these characteristics permit to identify a major origin located in the former region of the Gold Coast and the Bight of Benin; regions highly impacted by slavery, from which goes a sex-biased longitudinal gradient of ancestry.
Figures


References
-
- Klein HS. The Atlantic Slave Trade. Cambridge: Cambridge University Press; 1999.
-
- Eltis D, Behrendt SD, Richardson D, Klein HS. The Trans-Atlantic Slave Trade: A Database on CD-ROM. Cambridge; 1999.
-
- Price R, Price S. Les Marrons en Guyane. Châteauneuf-le-Rouge: Vents d'Ailleurs. 2002.
-
- Bortolini MC, Da Silva WAJW, De Guerra DC, Remonatto G, Mirandola R, Hutz MH, Weimer TA, Silva MC, Zago MA, Salzano FM. African-derived South American populations: A history of symmetrical and asymmetrical matings according to sex revealed by bi- and uni-parental genetic markers. Am J Hum Biol. 1999;11(4):551–563. doi: 10.1002/(SICI)1520-6300(1999)11:4<551::AID-AJHB15>3.0.CO;2-Z. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

