Interaction of Rep and DnaB on DNA
- PMID: 20959294
- PMCID: PMC3045612
- DOI: 10.1093/nar/gkq975
Interaction of Rep and DnaB on DNA
Abstract
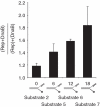
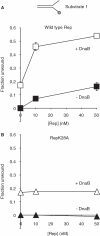
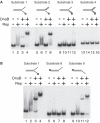
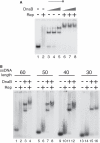
Genome duplication requires not only unwinding of the template but also the displacement of proteins bound to the template, a function performed by replicative helicases located at the fork. However, accessory helicases are also needed since the replicative helicase stalls occasionally at nucleoprotein complexes. In Escherichia coli, the primary and accessory helicases DnaB and Rep translocate along the lagging and leading strand templates, respectively, interact physically and also display cooperativity in the unwinding of model forked DNA substrates. We demonstrate here that this cooperativity is displayed only by Rep and not by other tested helicases. ssDNA must be exposed on the leading strand template to elicit this cooperativity, indicating that forks blocked at protein-DNA complexes contain ssDNA ahead of the leading strand polymerase. However, stable Rep-DnaB complexes can form on linear as well as branched DNA, indicating that Rep has the capacity to interact with ssDNA on either the leading or the lagging strand template at forks. Inhibition of Rep binding to the lagging strand template by competition with SSB might therefore be critical in targeting accessory helicases to the leading strand template, indicating an important role for replisome architecture in promoting accessory helicase function at blocked replisomes.
Figures



References
-
- French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992;258:1362–1365. - PubMed
-
- Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. - PubMed
-
- McGlynn P, Lloyd RG. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. - PubMed
-
- Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol. Cell. 2005;19:247–258. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

