Compensatory expression and substrate inducibility of gamma-glutamyl transferase GGT2 isoform in Arabidopsis thaliana
- PMID: 20959624
- PMCID: PMC3003821
- DOI: 10.1093/jxb/erq316
Compensatory expression and substrate inducibility of gamma-glutamyl transferase GGT2 isoform in Arabidopsis thaliana
Abstract
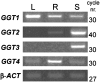
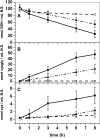
γ-Glutamyl transferases (GGT; EC 2.3.2.2) are glutathione-degrading enzymes that are represented in Arabidopsis thaliana by a small gene family of four members. Two isoforms, GGT1 and GGT2, are apoplastic, sharing broad similarities in their amino acid sequences, but they are differently expressed in the tissues: GGT1 is expressed in roots, leaves, and siliques, while GGT2 was thought to be expressed only in siliques. It is demonstrated here that GGT2 is also expressed in wild-type roots, albeit in very small amounts. GGT2 expression is enhanced in ggt1 knockout mutants, suggesting a compensatory effect to restore GGT activity in the root apoplast. Supplementation with 100 μM glutathione (GSH) resulted in the up-regulation of GGT2 gene expression in wild-type and ggt1 knockout roots, and of GGT1 gene expression in wild-type roots. Glutathione recovery was hampered by the GGT inhibitor serine/borate, suggesting a major role for apoplastic GGTs in this process. These findings can explain the ability of ggt1 knockout mutants to retrieve exogenously added glutathione from the growth medium.
Figures




Similar articles
-
Apoplastic gamma-glutamyl transferase activity encoded by GGT1 and GGT2 is important for vegetative and generative development.Plant Physiol Biochem. 2017 Jun;115:44-56. doi: 10.1016/j.plaphy.2017.03.007. Epub 2017 Mar 10. Plant Physiol Biochem. 2017. PMID: 28319794
-
Characterization of the extracellular gamma-glutamyl transpeptidases, GGT1 and GGT2, in Arabidopsis.Plant J. 2007 Mar;49(5):865-77. doi: 10.1111/j.1365-313X.2006.03004.x. Plant J. 2007. PMID: 17316175
-
Localization of members of the gamma-glutamyl transpeptidase family identifies sites of glutathione and glutathione S-conjugate hydrolysis.Plant Physiol. 2007 Aug;144(4):1715-32. doi: 10.1104/pp.106.094409. Epub 2007 Jun 1. Plant Physiol. 2007. PMID: 17545509 Free PMC article.
-
Biochemical and quantitative proteomics investigations in Arabidopsis ggt1 mutant leaves reveal a role for the gamma-glutamyl cycle in plant's adaptation to environment.Proteomics. 2013 Jun;13(12-13):2031-45. doi: 10.1002/pmic.201200479. Epub 2013 Jun 7. Proteomics. 2013. PMID: 23661340
-
gamma-Glutamyl transpeptidase: catalytic mechanism and gene expression.Adv Enzymol Relat Areas Mol Biol. 1998;72:239-78. doi: 10.1002/9780470123188.ch7. Adv Enzymol Relat Areas Mol Biol. 1998. PMID: 9559055 Review.
Cited by
-
Assigning gene function in biosynthetic pathways: camalexin and beyond.Plant Cell. 2013 Feb;25(2):360-7. doi: 10.1105/tpc.112.104745. Epub 2013 Feb 28. Plant Cell. 2013. PMID: 23449503 Free PMC article. No abstract available.
-
Glutathione Metabolism in Plants under Stress: Beyond Reactive Oxygen Species Detoxification.Metabolites. 2021 Sep 19;11(9):641. doi: 10.3390/metabo11090641. Metabolites. 2021. PMID: 34564457 Free PMC article. Review.
-
MdGGT1 Impacts Apple miR156 Precursor Levels via Ontogenetic Changes in Subcellular Glutathione Homeostasis.Front Plant Sci. 2019 Jul 31;10:994. doi: 10.3389/fpls.2019.00994. eCollection 2019. Front Plant Sci. 2019. PMID: 31417600 Free PMC article.
-
Glutathione degradation activity of γ-glutamyl peptidase 1 manifests its dual roles in primary and secondary sulfur metabolism in Arabidopsis.Plant J. 2022 Sep;111(6):1626-1642. doi: 10.1111/tpj.15912. Epub 2022 Aug 6. Plant J. 2022. PMID: 35932489 Free PMC article.
-
ROS-talk - how the apoplast, the chloroplast, and the nucleus get the message through.Front Plant Sci. 2012 Dec 27;3:292. doi: 10.3389/fpls.2012.00292. eCollection 2012. Front Plant Sci. 2012. PMID: 23293644 Free PMC article.
References
-
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genomewide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. - PubMed
-
- Beemster G, Fiorani F, Inzé D. Cell cycle: the key to plant growth control? Trends in Plant Science. 2003;8:154–158. - PubMed
-
- Dixon DP, Cummins I, Cole DJ, Edwards R. Glutathione-mediated detoxification systems in plants. Current Opinion in Plant Biology. 1998;1:258–266. - PubMed
-
- Edwards R, Dixon DP, Walbot V. Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends in Plant Science. 2000;5:193–198. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

