Molecular characterization of the evolution of phagosomes
- PMID: 20959821
- PMCID: PMC2990642
- DOI: 10.1038/msb.2010.80
Molecular characterization of the evolution of phagosomes
Abstract
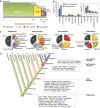
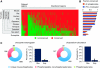
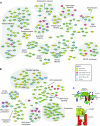
Amoeba use phagocytosis to internalize bacteria as a source of nutrients, whereas multicellular organisms utilize this process as a defense mechanism to kill microbes and, in vertebrates, initiate a sustained immune response. By using a large-scale approach to identify and compare the proteome and phosphoproteome of phagosomes isolated from distant organisms, and by comparative analysis over 39 taxa, we identified an 'ancient' core of phagosomal proteins around which the immune functions of this organelle have likely organized. Our data indicate that a larger proportion of the phagosome proteome, compared with the whole cell proteome, has been acquired through gene duplication at a period coinciding with the emergence of innate and adaptive immunity. Our study also characterizes in detail the acquisition of novel proteins and the significant remodeling of the phagosome phosphoproteome that contributed to modify the core constituents of this organelle in evolution. Our work thus provides the first thorough analysis of the changes that enabled the transformation of the phagosome from a phagotrophic compartment into an organelle fully competent for antigen presentation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Barabasi AL, Oltvai ZN (2004) Network biology: understanding the cell′s functional organization. Nat Rev Genet 5: 101–113 - PubMed
-
- Bertholet S, Goldszmid R, Morrot A, Debrabant A, Afrin F, Collazo-Custodio C, Houde M, Desjardins M, Sher A, Sacks D (2006) Leishmania antigens are presented to CD8+ T cells by a transporter associated with antigen processing-independent pathway in vitro and in vivo. J Immunol 177: 3525–3533 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

