POU homeodomain protein OCT1 modulates islet 1 expression during cardiac differentiation of P19CL6 cells
- PMID: 20960024
- PMCID: PMC11115038
- DOI: 10.1007/s00018-010-0544-y
POU homeodomain protein OCT1 modulates islet 1 expression during cardiac differentiation of P19CL6 cells
Abstract
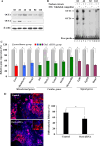
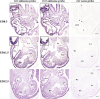
Islet 1 (ISL1), a marker of cardiac progenitors, plays a crucial role in cardiogenesis. However, the precise mechanism underlying the activation of its expression is not fully understood. Using the cardiac differentiation model of P19CL6 cells, we show that POU homeodomain protein, OCT1, modulates Isl1 expression in the process of cardiac differentiation. Oct1 knock-down resulted in reduction of Isl1 expression and downregulated mesodermal, cardiac-specific, and signal pathway gene expression. Additionally, the octamer motif located in the proximal region of Isl1 promoter is essential to Isl1 transcriptional activation. Mutation of this motif remarkably decreased Isl1 transcription. Although both OCT1 and OCT4 bound to this motif, it was OCT1 rather than OCT4 that modulated Isl1 expression. Furthermore, the correlation of OCT1 in regulation of Isl1 was revealed by in situ hybridization in early embryos. Collectively, our data highlight a novel role of OCT1 in the regulation of Isl1 expression.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

