Cytokine and progesterone receptor interplay in the regulation of MUC1 gene expression
- PMID: 20962044
- PMCID: PMC2999481
- DOI: 10.1210/me.2009-0448
Cytokine and progesterone receptor interplay in the regulation of MUC1 gene expression
Abstract
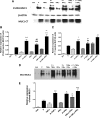
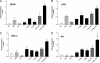
Mucin 1 (MUC1), a transmembrane mucin expressed at the apical surface of uterine epithelia, is a barrier to microbial infection and enzymatic attack. MUC1 loss at implantation sites appears to be required to permit embryo attachment and implantation in most species. MUC1 expression is regulated by progesterone (P) and proinflammatory cytokines, including TNFα and interferon γ (IFNγ). TNFα and IFNγ are highly expressed in uterine tissues under conditions where MUC1 expression is also high and activate MUC1 expression via their downstream transcription factors, nuclear factor (NF) κB and signal transducers and activators of transcription. P receptor (PR) regulates MUC1 gene expression in a PR isoform-specific fashion. Here we demonstrate that interactions among PR isoforms and cytokine-activated transcription factors cooperatively regulate MUC1 expression in a human uterine epithelial cell line, HES. Low doses of IFNγ and TNFα synergistically stimulate MUC1 promoter activity, enhance PRB stimulation of MUC1 promoter activity and cooperate with PRA to stimulate MUC1 promoter activity. Cooperative stimulation of MUC1 promoter activity requires the DNA-binding domain of the PR isoforms. MUC1 mRNA and protein expression is increased by cytokine and P treatment in HES cells stably expressing PRB. Using chromatin immunoprecipitation assays, we demonstrate efficient recruitment of NFκB, p300, SRC3 (steroid receptor coactivator 3), and PR to the MUC1 promoter. Collectively, our studies indicate a dynamic interplay among cytokine-activated transcription factors, PR isoforms and transcriptional coregulators in modulating MUC1 expression. This interplay may have important consequences in both normal and pathological contexts, e.g. implantation failure and recurrent miscarriages.
Figures


Similar articles
-
Progesterone receptor isoforms A and B differentially regulate MUC1 expression in uterine epithelial cells.Mol Endocrinol. 2006 Oct;20(10):2278-91. doi: 10.1210/me.2005-0343. Epub 2006 Jun 1. Mol Endocrinol. 2006. PMID: 16740655
-
MUC1 expression is repressed by protein inhibitor of activated signal transducer and activator of transcription-y.Mol Endocrinol. 2007 Nov;21(11):2725-37. doi: 10.1210/me.2006-0539. Epub 2007 Aug 23. Mol Endocrinol. 2007. PMID: 17717071
-
Synergistic stimulation of MUC1 expression in normal breast epithelia and breast cancer cells by interferon-gamma and tumor necrosis factor-alpha.J Cell Biochem. 2002;86(4):759-72. doi: 10.1002/jcb.10261. J Cell Biochem. 2002. PMID: 12210742
-
Role of nuclear progesterone receptor isoforms in uterine pathophysiology.Hum Reprod Update. 2015 Mar-Apr;21(2):155-73. doi: 10.1093/humupd/dmu056. Epub 2014 Nov 18. Hum Reprod Update. 2015. PMID: 25406186 Free PMC article. Review.
-
More help than hindrance: nucleosomes aid transcriptional regulation.Nucleus. 2013 May-Jun;4(3):189-94. doi: 10.4161/nucl.25108. Epub 2013 May 28. Nucleus. 2013. PMID: 23756349 Free PMC article. Review.
Cited by
-
Discovery of HeLa Cell Contamination in HES Cells: Call for Cell Line Authentication in Reproductive Biology Research.Reprod Sci. 2014 Aug;21(8):1015-1019. doi: 10.1177/1933719114522518. Epub 2014 Feb 11. Reprod Sci. 2014. PMID: 24520087 Free PMC article.
-
Pasteurella multocida specific bacteriophage suppresses P. multocida-induced inflammation: identification of genes related to bacteriophage signaling by Pasteurella multocida-infected swine nasal turbinate cells.Genes Genomics. 2020 Feb;42(2):235-243. doi: 10.1007/s13258-019-00898-4. Epub 2019 Dec 18. Genes Genomics. 2020. PMID: 31853889
-
The role of inflammation for a successful implantation.Am J Reprod Immunol. 2014 Aug;72(2):141-7. doi: 10.1111/aji.12266. Epub 2014 May 9. Am J Reprod Immunol. 2014. PMID: 24809430 Free PMC article. Review.
-
Endometrial receptivity defects and impaired implantation in diabetic NOD mice.Biol Reprod. 2012 Aug 2;87(2):30. doi: 10.1095/biolreprod.112.100016. Print 2012 Aug. Biol Reprod. 2012. PMID: 22539679 Free PMC article.
-
MUC1 (CD227): a multi-tasked molecule.Cell Mol Life Sci. 2015 Dec;72(23):4475-500. doi: 10.1007/s00018-015-2014-z. Epub 2015 Aug 21. Cell Mol Life Sci. 2015. PMID: 26294353 Free PMC article. Review.
References
-
- Gendler SJ 2001 MUC1, the renaissance molecule. J Mammary Gland Biol Neoplasia 6:339–353 - PubMed
-
- Gendler SJ, Spicer AP 1995 Epithelial mucin genes. Annu Rev Physiol 57:607–634 - PubMed
-
- Thathiah A, Carson DD 2002 Mucins and blastocyst attachment. Rev Endocr Metab Disord 3:87–96 - PubMed
-
- Lagow E, DeSouza MM, Carson DD 1999 Mammalian reproductive tract mucins. Hum Reprod Update 5:280–292 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

