Simian immunodeficiency virus from the sooty mangabey and rhesus macaque is modified with O-linked carbohydrate
- PMID: 20962077
- PMCID: PMC3014205
- DOI: 10.1128/JVI.01871-10
Simian immunodeficiency virus from the sooty mangabey and rhesus macaque is modified with O-linked carbohydrate
Abstract
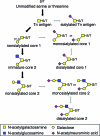
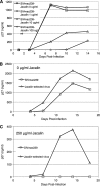
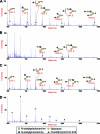
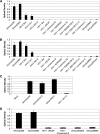
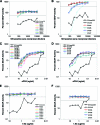
Although stretches of serine and threonine are sometimes sites for O-linked carbohydrate attachment, specific sequence and structural determinants for O-linked attachment remain ill defined. The gp120 envelope protein of SIVmac239 contains a serine-threonine-rich stretch of amino acids at positions 128 to 139. Here we show that lectin protein from jackfruit seed (jacalin), which binds to non- and monosialylated core 1 O-linked carbohydrate, potently inhibited the replication of SIVmac239. Selection of a jacalin-resistant SIVmac239 variant population resulted in virus with specific substitutions within amino acids 128 to 139. Cloned simian immunodeficiency virus (SIV) variants with substitutions in the 128-to-139 region had infectivities equivalent to, or within 1 log unit of, that of SIVmac239 and were resistant to the inhibitory effects of jacalin. Characterization of the SIVmac239 gp120 O-linked glycome showed the presence of core 1 and core 2 O-linked carbohydrate; a 128-to-139-substituted variant gp120 from jacalin-resistant SIV lacked O-linked carbohydrate. Unlike that of SIVmac239, the replication of HIV-1 strain NL4-3 was resistant to inhibition by jacalin. Purified gp120s from four SIVmac and SIVsm strains bound jacalin strongly in an enzyme-linked immunosorbent assay, while nine different HIV-1 gp120s, two SIVcpz gp120s, and 128-to-139-substituted SIVmac239 gp120 did not bind jacalin. The ability or inability to bind jacalin thus correlated with the presence of the serine-threonine-rich stretch in the SIVmac and SIVsm gp120s and the absence of such stretches in the SIVcpz and HIV-1 gp120s. Consistent with sequence predictions, two HIV-2 gp120s bound jacalin, while one did not. These data demonstrate the presence of non- and monosialylated core 1 O-linked carbohydrate on the gp120s of SIVmac and SIVsm and the lack of these modifications on HIV-1 and SIVcpz gp120s.
Figures




Similar articles
-
Fundamental difference in the content of high-mannose carbohydrate in the HIV-1 and HIV-2 lineages.J Virol. 2010 Sep;84(18):8998-9009. doi: 10.1128/JVI.00996-10. Epub 2010 Jul 7. J Virol. 2010. PMID: 20610711 Free PMC article.
-
Primary SIVsm isolates use the CCR5 coreceptor from sooty mangabeys naturally infected in west Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2, and SIVmac.Virology. 1998 Jun 20;246(1):113-24. doi: 10.1006/viro.1998.9174. Virology. 1998. PMID: 9656999
-
Human Immunodeficiency Virus and Simian Immunodeficiency Virus Maintain High Levels of Infectivity in the Complete Absence of Mucin-Type O-Glycosylation.J Virol. 2017 Sep 12;91(19):e01228-17. doi: 10.1128/JVI.01228-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28747495 Free PMC article.
-
Primate models for human immunodeficiency virus infection. Evolution of receptor use during pathogenesis.Acta Microbiol Immunol Hung. 2004;51(1-2):1-29. doi: 10.1556/AMicr.51.2004.1-2.1. Acta Microbiol Immunol Hung. 2004. PMID: 15362285 Review.
-
The function of simian chemokine receptors in the replication of SIV.Semin Immunol. 1998 Jun;10(3):215-23. doi: 10.1006/smim.1998.0135. Semin Immunol. 1998. PMID: 9653048 Review.
Cited by
-
A recombinant herpesviral vector containing a near-full-length SIVmac239 genome produces SIV particles and elicits immune responses to all nine SIV gene products.PLoS Pathog. 2018 Jun 18;14(6):e1007143. doi: 10.1371/journal.ppat.1007143. eCollection 2018 Jun. PLoS Pathog. 2018. PMID: 29912986 Free PMC article.
-
HEK293T cell lines defective for O-linked glycosylation.PLoS One. 2017 Jun 27;12(6):e0179949. doi: 10.1371/journal.pone.0179949. eCollection 2017. PLoS One. 2017. PMID: 28654657 Free PMC article.
-
Characterization of the viral O-glycopeptidome: a novel tool of relevance for vaccine design and serodiagnosis.J Virol. 2012 Jun;86(11):6268-78. doi: 10.1128/JVI.00392-12. Epub 2012 Apr 4. J Virol. 2012. PMID: 22491453 Free PMC article.
-
High-Resolution Sequencing of Viral Populations during Early Simian Immunodeficiency Virus Infection Reveals Evolutionary Strategies for Rapid Escape from Emerging Env-Specific Antibody Responses.J Virol. 2018 Mar 14;92(7):e01574-17. doi: 10.1128/JVI.01574-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29343575 Free PMC article.
-
Comparative Analysis of the Glycosylation Profiles of Membrane-Anchored HIV-1 Envelope Glycoprotein Trimers and Soluble gp140.J Virol. 2015 Aug;89(16):8245-57. doi: 10.1128/JVI.00628-15. Epub 2015 May 27. J Virol. 2015. PMID: 26018173 Free PMC article.
References
-
- Baba, M., B. Yong Ma, M. Nonaka, Y. Matsuishi, M. Hirano, N. Nakamura, N. Kawasaki, and T. Kawasaki. 2007. Glycosylation-dependent interaction of jacalin with CD45 induces T lymphocyte activation and Th1/Th2 cytokine secretion. J. Leukoc. Biol. 81:1002-1011. - PubMed
-
- Ceroni, A., K. Maass, H. Geyer, R. Geyer, A. Dell, and S. M. Haslam. 2008. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7:1650-1659. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

