Role of RNase MRP in viral RNA degradation and RNA recombination
- PMID: 20962095
- PMCID: PMC3014185
- DOI: 10.1128/JVI.01749-10
Role of RNase MRP in viral RNA degradation and RNA recombination
Abstract
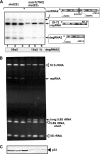
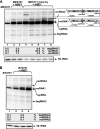
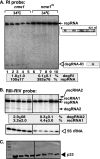
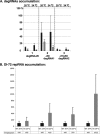
RNA degradation, together with RNA synthesis, controls the steady-state level of viral RNAs in infected cells. The endoribonucleolytic cleavage of viral RNA is important not only for viral RNA degradation but for RNA recombination as well, due to the participation of some RNA degradation products in the RNA recombination process. To identify host endoribonucleases involved in degradation of Tomato bushy stunt virus (TBSV) in a Saccharomyces cerevisiae model host, we tested eight known endoribonucleases. Here we report that downregulation of SNM1, encoding a component of the RNase MRP, and a temperature-sensitive mutation in the NME1 gene, coding for the RNA component of RNase MRP, lead to reduced production of the endoribonucleolytically cleaved TBSV RNA in yeast. We also show that the highly purified yeast RNase MRP cleaves the TBSV RNA in vitro, resulting in TBSV RNA degradation products similar in size to those observed in yeast cells. Knocking down the NME1 homolog in Nicotiana benthamiana also led to decreased production of the cleaved TBSV RNA, suggesting that in plants, RNase MRP is involved in TBSV RNA degradation. Altogether, this work suggests a role for the host endoribonuclease RNase MRP in viral RNA degradation and recombination.
Figures




Similar articles
-
Screening bacterial effectors and human virus proteins in yeast to identify host factors driving tombusvirus RNA recombination: a role for autophagy and membrane phospholipid content.J Virol. 2025 Jun 17;99(6):e0166124. doi: 10.1128/jvi.01661-24. Epub 2025 May 27. J Virol. 2025. PMID: 40422074 Free PMC article.
-
Co-opted Cellular Sac1 Lipid Phosphatase and PI(4)P Phosphoinositide Are Key Host Factors during the Biogenesis of the Tombusvirus Replication Compartment.J Virol. 2020 Jun 1;94(12):e01979-19. doi: 10.1128/JVI.01979-19. Print 2020 Jun 1. J Virol. 2020. PMID: 32269127 Free PMC article.
-
Interviral Recombination between Plant, Insect, and Fungal RNA Viruses: Role of the Intracellular Ca2+/Mn2+ Pump.J Virol. 2019 Dec 12;94(1):e01015-19. doi: 10.1128/JVI.01015-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597780 Free PMC article.
-
The roles of host factors in tombusvirus RNA recombination.Adv Virus Res. 2011;81:63-84. doi: 10.1016/B978-0-12-385885-6.00008-0. Adv Virus Res. 2011. PMID: 22094079 Review.
-
Genetic and biochemical analyses of yeast RNase MRP.Mol Biol Rep. 1995-1996;22(2-3):75-9. doi: 10.1007/BF00988709. Mol Biol Rep. 1995. PMID: 8901491 Review.
Cited by
-
Modulation of neuronal proteome profile in response to Japanese encephalitis virus infection.PLoS One. 2014 Mar 5;9(3):e90211. doi: 10.1371/journal.pone.0090211. eCollection 2014. PLoS One. 2014. PMID: 24599148 Free PMC article.
-
Authentic in vitro replication of two tombusviruses in isolated mitochondrial and endoplasmic reticulum membranes.J Virol. 2012 Dec;86(23):12779-94. doi: 10.1128/JVI.00973-12. Epub 2012 Sep 12. J Virol. 2012. PMID: 22973028 Free PMC article.
-
Footprinting analysis of interactions between the largest eukaryotic RNase P/MRP protein Pop1 and RNase P/MRP RNA components.RNA. 2015 Sep;21(9):1591-605. doi: 10.1261/rna.049007.114. Epub 2015 Jul 1. RNA. 2015. PMID: 26135751 Free PMC article.
-
SARS-CoV-2 Subgenomic RNAs: Characterization, Utility, and Perspectives.Viruses. 2021 Sep 24;13(10):1923. doi: 10.3390/v13101923. Viruses. 2021. PMID: 34696353 Free PMC article. Review.
-
RNA structural elements determine frequency and sites of nonhomologous recombination in an animal plus-strand RNA virus.J Virol. 2012 Jul;86(13):7393-402. doi: 10.1128/JVI.00864-12. Epub 2012 Apr 24. J Virol. 2012. PMID: 22532677 Free PMC article.
References
-
- Aaziz, R., and M. Tepfer. 1999. Recombination in RNA viruses and in virus-resistant transgenic plants. J. Gen. Virol. 80:1339-1346. - PubMed
-
- Cai, T., and M. E. Schmitt. 2001. Characterization of ribonuclease MRP function. Methods Enzymol. 342:135-142. - PubMed
-
- Cheng, C. P., H. M. Jaag, M. Jonczyk, E. Serviene, and P. D. Nagy. 2007. Expression of the Arabidopsis Xrn4p 5′-3′ exoribonuclease facilitates degradation of tombusvirus RNA and promotes rapid emergence of viral variants in plants. Virology 368:238-248. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

