Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice
- PMID: 20962874
- PMCID: PMC3105739
- DOI: 10.1038/ismej.2010.162
Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice
Abstract
This study is the first to use a metagenomics approach to characterize the phylogeny and functional capacity of the canine gastrointestinal microbiome. Six healthy adult dogs were used in a crossover design and fed a low-fiber control diet (K9C) or one containing 7.5% beet pulp (K9BP). Pooled fecal DNA samples from each treatment were subjected to 454 pyrosequencing, generating 503,280 (K9C) and 505,061 (K9BP) sequences. Dominant bacterial phyla included the Bacteroidetes/Chlorobi group and Firmicutes, both of which comprised ∼35% of all sequences, followed by Proteobacteria (13-15%) and Fusobacteria (7-8%). K9C had a greater percentage of Bacteroidetes, Fusobacteria and Proteobacteria, whereas K9BP had greater proportions of the Bacteroidetes/Chlorobi group and Firmicutes. Archaea were not altered by diet and represented ∼1% of all sequences. All archaea were members of Crenarchaeota and Euryarchaeota, with methanogens being the most abundant and diverse. Three fungi phylotypes were present in K9C, but none in K9BP. Less than 0.4% of sequences were of viral origin, with >99% of them associated with bacteriophages. Primary functional categories were not significantly affected by diet and were associated with carbohydrates; protein metabolism; DNA metabolism; cofactors, vitamins, prosthetic groups and pigments; amino acids and derivatives; cell wall and capsule; and virulence. Hierarchical clustering of several gastrointestinal metagenomes demonstrated phylogenetic and metabolic similarity between dogs, humans and mice. More research is required to provide deeper coverage of the canine microbiome, evaluate effects of age, genetics or environment on its composition and activity, and identify its role in gastrointestinal disease.
Figures
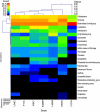
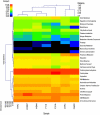
References
-
- Abnous K, Brooks SPJ, Kwan J, Matias F, Green-Johnson J, Selinger LB, et al. Diets enriched in oat bran or wheat bran temporally and differentially alter the composition of the fecal community of rats. J Nutr. 2009;139:2024–2031. - PubMed
-
- Allenspach K, House A, Smith K, McNeill FM, Hendricks A, Elson-Riggins J, et al. 2010Evaluation of mucosal bacteria and histopathology, clinical disease activity and expression of Toll-like receptors in German shepherd dogs with chronic enteropathies Vet Microbioldoi: 10.1016/j.vetmic.2010.05.025 - DOI - PubMed
-
- Alonso G, Vilchez G, Lemoine VR. How bacteria protect themselves against channel-forming colicins. Int Microbiol. 2000;3:81–88. - PubMed
-
- Association of American Feed Control Officials . Official Publication of the Association of American Feed Control Officials. Association of American Feed Control Officials, Inc: Oxford, IN, USA; 2009.
-
- Benno Y, Nakao H, Uchida K, Mitsuoka T. Impact of the advances in age on the gastrointestinal microflora of beagle dogs. J Vet Med Sci. 1992;54:703–706. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

