Capturing diversity of marine heterotrophic protists: one cell at a time
- PMID: 20962875
- PMCID: PMC3105736
- DOI: 10.1038/ismej.2010.155
Capturing diversity of marine heterotrophic protists: one cell at a time
Abstract
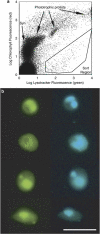
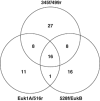
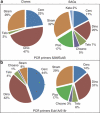
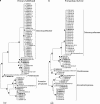
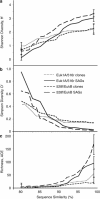
Recent applications of culture-independent, molecular methods have revealed unexpectedly high diversity in a variety of functional and phylogenetic groups of microorganisms in the ocean. However, none of the existing research tools are free from significant limitations, such as PCR and cloning biases, low phylogenetic resolution and others. Here, we employed novel, single-cell sequencing techniques to assess the composition of small (<10 μm diameter), heterotrophic protists from the Gulf of Maine. Single cells were isolated by flow cytometry, their genomes amplified, and 18S rRNA marker genes were amplified and sequenced. We compared the results to traditional environmental PCR cloning of sorted cells. The diversity of heterotrophic protists was significantly higher in the library of single amplified genomes (SAGs) than in environmental PCR clone libraries of the 18S rRNA gene, obtained from the same coastal sample. Libraries of SAGs, but not clones contained several recently discovered, uncultured groups, including picobiliphytes and novel marine stramenopiles. Clone, but not SAG, libraries contained several large clusters of identical and nearly identical sequences of Dinophyceae, Cercozoa and Stramenopiles. Similar results were obtained using two alternative primer sets, suggesting that PCR biases may not be the only explanation for the observed patterns. Instead, differences in the number of 18S rRNA gene copies among the various protist taxa probably had a significant role in determining the PCR clone composition. These results show that single-cell sequencing has the potential to more accurately assess protistan community composition than previously established methods. In addition, the creation of SAG libraries opens opportunities for the analysis of multiple genes or entire genomes of the uncultured protist groups.
Figures
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Azam F, Fenchel T, Field JG, Meyer-Reil LA, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Progr Ser. 1983;10:257–263.

