Selection of Shine-Dalgarno sequences in plastids
- PMID: 20965967
- PMCID: PMC3045613
- DOI: 10.1093/nar/gkq978
Selection of Shine-Dalgarno sequences in plastids
Abstract
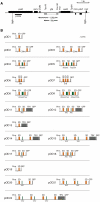
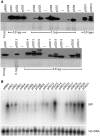
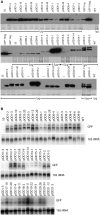
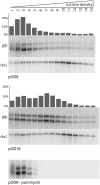
Like bacterial genes, most plastid (chloroplast) genes are arranged in operons and transcribed as polycistronic mRNAs. Plastid protein biosynthesis occurs on bacterial-type 70S ribosomes and translation initiation of many (but not all) mRNAs is mediated by Shine-Dalgarno (SD) sequences. To study the mechanisms of SD sequence recognition, we have analyzed translation initiation from mRNAs containing multiple SD sequences. Comparing translational efficiencies of identical transgenic mRNAs in Escherichia coli and plastids, we find surprising differences between the two systems. Most importantly, while internal SD sequences are efficiently recognized in E. coli, plastids exhibit a bias toward utilizing predominantly the 5'-most SD sequence. We propose that inefficient recognition of internal SD sequences provides the raison d'être for most plastid polycistronic transcripts undergoing post-transcriptional cleavage into monocistronic mRNAs.
Figures
References
-
- Liere K, Börner T. Transcription and transcriptional regulation in plastids. Top. Curr. Genet. 2007;19:121–174.
-
- Sugiura M, Hirose T, Sugita M. Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet. 1998;32:437–459. - PubMed
-
- Beligni MV, Yamaguchi K, Mayfield SP. The translational apparatus of Chlamydomonas reinhardtii chloroplast. Photosynth. Res. 2004;82:315–325. - PubMed
-
- Peled-Zehavi H, Danon A. Translation and translational regulation in chloroplasts. Top. Curr. Genet. 2007;19:249–281.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

