Microhomology-mediated deletion and gene conversion in African trypanosomes
- PMID: 20965968
- PMCID: PMC3045614
- DOI: 10.1093/nar/gkq981
Microhomology-mediated deletion and gene conversion in African trypanosomes
Abstract
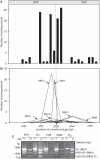
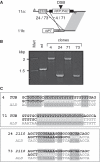
Antigenic variation in African trypanosomes is induced by DNA double-strand breaks (DSBs). In these protozoan parasites, DSB repair (DSBR) is dominated by homologous recombination (HR) and microhomology-mediated end joining (MMEJ), while non-homologous end joining (NHEJ) has not been reported. To facilitate the analysis of chromosomal end-joining, we established a system whereby inter-allelic repair by HR is lethal due to loss of an essential gene. Analysis of intrachromosomal end joining in individual DSBR survivors exclusively revealed MMEJ-based deletions but no NHEJ. A survey of microhomologies typically revealed sequences of between 5 and 20 bp in length with several mismatches tolerated in longer stretches. Mean deletions were of 54 bp on the side closest to the break and 284 bp in total. Break proximity, microhomology length and GC-content all favored repair and the pattern of MMEJ described above was similar at several different loci across the genome. We also identified interchromosomal gene conversion involving HR and MMEJ at different ends of a duplicated sequence. While MMEJ-based deletions were RAD51-independent, one-sided MMEJ was RAD51 dependent. Thus, we describe the features of MMEJ in Trypanosoma brucei, which is analogous to micro single-strand annealing; and RAD51 dependent, one-sided MMEJ. We discuss the contribution of MMEJ pathways to genome evolution, subtelomere recombination and antigenic variation.
Figures




Similar articles
-
Sequence homology and microhomology dominate chromosomal double-strand break repair in African trypanosomes.Nucleic Acids Res. 2008 May;36(8):2608-18. doi: 10.1093/nar/gkn104. Epub 2008 Mar 11. Nucleic Acids Res. 2008. PMID: 18334531 Free PMC article.
-
Single-Strand Annealing Plays a Major Role in Double-Strand DNA Break Repair following CRISPR-Cas9 Cleavage in Leishmania.mSphere. 2019 Aug 21;4(4):e00408-19. doi: 10.1128/mSphere.00408-19. mSphere. 2019. PMID: 31434745 Free PMC article.
-
Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination.Genetics. 2007 Jul;176(3):1403-15. doi: 10.1534/genetics.107.071621. Epub 2007 May 4. Genetics. 2007. PMID: 17483423 Free PMC article.
-
Microhomology-mediated end joining: Good, bad and ugly.Mutat Res. 2018 May;809:81-87. doi: 10.1016/j.mrfmmm.2017.07.002. Epub 2017 Jul 16. Mutat Res. 2018. PMID: 28754468 Free PMC article. Review.
-
Contribution of Microhomology to Genome Instability: Connection between DNA Repair and Replication Stress.Int J Mol Sci. 2022 Oct 26;23(21):12937. doi: 10.3390/ijms232112937. Int J Mol Sci. 2022. PMID: 36361724 Free PMC article. Review.
Cited by
-
Malaria parasites utilize both homologous recombination and alternative end joining pathways to maintain genome integrity.Nucleic Acids Res. 2014 Jan;42(1):370-9. doi: 10.1093/nar/gkt881. Epub 2013 Oct 1. Nucleic Acids Res. 2014. PMID: 24089143 Free PMC article.
-
Keeping Balance Between Genetic Stability and Plasticity at the Telomere and Subtelomere of Trypanosoma brucei.Front Cell Dev Biol. 2021 Jul 5;9:699639. doi: 10.3389/fcell.2021.699639. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34291053 Free PMC article. Review.
-
A Naturally Occurring Microhomology-Mediated Deletion of Three Genes in African Swine Fever Virus Isolated from Two Sardinian Wild Boars.Viruses. 2022 Nov 14;14(11):2524. doi: 10.3390/v14112524. Viruses. 2022. PMID: 36423133 Free PMC article.
-
Trypanosomal histone γH2A and the DNA damage response.Mol Biochem Parasitol. 2012 May;183(1):78-83. doi: 10.1016/j.molbiopara.2012.01.008. Epub 2012 Feb 14. Mol Biochem Parasitol. 2012. PMID: 22353557 Free PMC article.
-
Efficient Single-Gene and Gene Family Editing in the Apicomplexan Parasite Eimeria tenella Using CRISPR-Cas9.Front Bioeng Biotechnol. 2020 Feb 25;8:128. doi: 10.3389/fbioe.2020.00128. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32158750 Free PMC article.
References
-
- Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. - PubMed
-
- Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. - PubMed
-
- Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131:223–225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

