Common and divergent features in transcriptional control of the homologous small RNAs GlmY and GlmZ in Enterobacteriaceae
- PMID: 20965974
- PMCID: PMC3045617
- DOI: 10.1093/nar/gkq986
Common and divergent features in transcriptional control of the homologous small RNAs GlmY and GlmZ in Enterobacteriaceae
Abstract
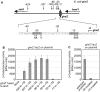
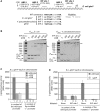
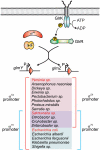
Small RNAs GlmY and GlmZ compose a cascade that feedback-regulates synthesis of enzyme GlmS in Enterobacteriaceae. Here, we analyzed the transcriptional regulation of glmY/glmZ from Yersinia pseudotuberculosis, Salmonella typhimurium and Escherichia coli, as representatives for other enterobacterial species, which exhibit similar promoter architectures. The GlmY and GlmZ sRNAs of Y. pseudotuberculosis are transcribed from σ(54)-promoters that require activation by the response regulator GlrR through binding to three conserved sites located upstream of the promoters. This also applies to glmY/glmZ of S. typhimurium and glmY of E. coli, but as a difference additional σ(70)-promoters overlap the σ(54)-promoters and initiate transcription at the same site. In contrast, E. coli glmZ is transcribed from a single σ(70)-promoter. Thus, transcription of glmY and glmZ is controlled by σ(54) and the two-component system GlrR/GlrK (QseF/QseE) in Y. pseudotuberculosis and presumably in many other Enterobacteria. However, in a subset of species such as E. coli this relationship is partially lost in favor of σ(70)-dependent transcription. In addition, we show that activity of the σ(54)-promoter of E. coli glmY requires binding of the integration host factor to sites upstream of the promoter. Finally, evidence is provided that phosphorylation of GlrR increases its activity and thereby sRNA expression.
Figures
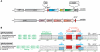




Similar articles
-
Dual control by perfectly overlapping sigma 54- and sigma 70- promoters adjusts small RNA GlmY expression to different environmental signals.Mol Microbiol. 2009 Dec;74(5):1054-70. doi: 10.1111/j.1365-2958.2009.06918.x. Epub 2009 Oct 27. Mol Microbiol. 2009. PMID: 19843219
-
Ménage à trois: post-transcriptional control of the key enzyme for cell envelope synthesis by a base-pairing small RNA, an RNase adaptor protein, and a small RNA mimic.RNA Biol. 2014;11(5):433-42. doi: 10.4161/rna.28301. Epub 2014 Feb 27. RNA Biol. 2014. PMID: 24667238 Free PMC article. Review.
-
Small RNA-binding protein RapZ mediates cell envelope precursor sensing and signaling in Escherichia coli.EMBO J. 2020 Mar 16;39(6):e103848. doi: 10.15252/embj.2019103848. Epub 2020 Feb 17. EMBO J. 2020. PMID: 32065419 Free PMC article.
-
The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli.Nucleic Acids Res. 2008 May;36(8):2570-80. doi: 10.1093/nar/gkn091. Epub 2008 Mar 11. Nucleic Acids Res. 2008. PMID: 18334534 Free PMC article.
-
Regulated Control of the Assembly and Diversity of LPS by Noncoding sRNAs.Biomed Res Int. 2015;2015:153561. doi: 10.1155/2015/153561. Epub 2015 Nov 5. Biomed Res Int. 2015. PMID: 26618164 Free PMC article. Review.
Cited by
-
Origin, Evolution, and Loss of Bacterial Small RNAs.Microbiol Spectr. 2018 Apr;6(2):10.1128/microbiolspec.rwr-0004-2017. doi: 10.1128/microbiolspec.RWR-0004-2017. Microbiol Spectr. 2018. PMID: 29623872 Free PMC article. Review.
-
Uptake and metabolism of N-acetylglucosamine and glucosamine by Streptococcus mutans.Appl Environ Microbiol. 2014 Aug;80(16):5053-67. doi: 10.1128/AEM.00820-14. Epub 2014 Jun 13. Appl Environ Microbiol. 2014. PMID: 24928869 Free PMC article.
-
GlmS and NagB regulate amino sugar metabolism in opposing directions and affect Streptococcus mutans virulence.PLoS One. 2012;7(3):e33382. doi: 10.1371/journal.pone.0033382. Epub 2012 Mar 16. PLoS One. 2012. PMID: 22438919 Free PMC article.
-
NagR Differentially Regulates the Expression of the glmS and nagAB Genes Required for Amino Sugar Metabolism by Streptococcus mutans.J Bacteriol. 2015 Nov;197(22):3533-44. doi: 10.1128/JB.00606-15. Epub 2015 Aug 31. J Bacteriol. 2015. PMID: 26324448 Free PMC article.
-
Bacterial Chat: Intestinal Metabolites and Signals in Host-Microbiota-Pathogen Interactions.Infect Immun. 2017 Nov 17;85(12):e00476-17. doi: 10.1128/IAI.00476-17. Print 2017 Dec. Infect Immun. 2017. PMID: 28947641 Free PMC article. Review.
References
-
- Görke B, Vogel J. Noncoding RNA control of the making and breaking of sugars. Genes Dev. 2008;22:2914–2925. - PubMed
-
- Repoila F, Darfeuille F. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol. Cell. 2009;101:117–131. - PubMed
-
- Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. - PubMed
-
- Brantl S. Bacterial chromosome-encoded small regulatory RNAs. Future Microbiol. 2009;4:85–103. - PubMed
-
- Vanderpool CK. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr. Opin. Microbiol. 2007;10:146–151. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

