Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila
- PMID: 20966049
- PMCID: PMC2975926
- DOI: 10.1101/gad.1968110
Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila
Abstract
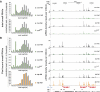
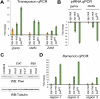
Combining RNAi in cultured cells and analysis of mutant animals, we probed the roles of known Piwi-interacting RNA (piRNA) pathway components in the initiation and effector phases of transposon silencing. Squash associated physically with Piwi, and reductions in its expression led to modest transposon derepression without effects on piRNAs, consistent with an effector role. Alterations in Zucchini or Armitage reduced both Piwi protein and piRNAs, indicating functions in the formation of a stable Piwi RISC (RNA-induced silencing complex). Notably, loss of Zucchini or mutations within its catalytic domain led to accumulation of unprocessed precursor transcripts from flamenco, consistent with a role for this putative nuclease in piRNA biogenesis.
Figures



References
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 - PubMed
-
- Aravin AA, Hannon GJ, Brennecke J 2007. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 - PubMed
-
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
