Functional dissection of adenylate cyclase R, an inducer of spore encapsulation
- PMID: 20966074
- PMCID: PMC3009899
- DOI: 10.1074/jbc.M110.156380
Functional dissection of adenylate cyclase R, an inducer of spore encapsulation
Abstract
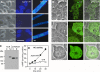
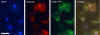
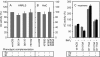
Cyclic AMP acting on protein kinase A controls sporulation and encystation in social and solitary amoebas. In Dictyostelium discoideum, adenylate cyclase R (ACR), is essential for spore encapsulation. In addition to its cyclase (AC) domain, ACR harbors seven transmembrane helices, a histidine kinase domain, and two receiver domains. We investigated the role of these domains in the regulation of AC activity. Expression of an ACR-YFP fusion protein in acr(-) cells rescued their sporulation defective phenotype and revealed that ACR is associated with the nuclear envelope and endoplasmic reticulum. Loss of the transmembrane helices (ΔTM) caused a 60% reduction of AC activity, but ΔTM-ACR still rescued the acr(-) phenotype. The isolated AC domain was properly expressed but inactive. Mutation of three essential ATP-binding residues in the histidine kinase domain did not affect the AC activity or phenotypic rescue. Mutation of the essential phosphoryl-accepting aspartate in receivers 1, 2, or both had only modest effects on AC activity and did not affect phenotypic rescue, indicating that AC activity is not critically regulated by phosphorelay. Remarkably, the dimerizing histidine phosphoacceptor subdomain, which in ACR lacks the canonical histidine for autophosphorylation, was essential for AC activity. Transformation of wild-type cells with an ACR allele (ΔCRA) that is truncated after this domain inhibited AC activity of endogenous ACR and replicated the acr(-) phenotype. Combined with the observation that the isolated AC domain was inactive, the dominant-negative effect of ΔCRA strongly suggests that the defunct phosphoacceptor domain acquired a novel role in enforcing dimerization of the AC domain.
Figures

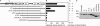
Similar articles
-
An adenylyl cyclase that functions during late development of Dictyostelium.Development. 1999 Dec;126(23):5463-71. doi: 10.1242/dev.126.23.5463. Development. 1999. PMID: 10556070
-
Dissection of the functional and structural domains of phosphorelay histidine kinase A of Bacillus subtilis.J Bacteriol. 2001 May;183(9):2795-802. doi: 10.1128/JB.183.9.2795-2802.2001. J Bacteriol. 2001. PMID: 11292798 Free PMC article.
-
Guanylate cyclase in Dictyostelium discoideum with the topology of mammalian adenylate cyclase.Biochem J. 2001 Mar 15;354(Pt 3):697-706. doi: 10.1042/0264-6021:3540697. Biochem J. 2001. PMID: 11237875 Free PMC article.
-
Ege A, a novel C2 domain containing protein, is essential for GPCR-mediated gene expression in dictyostelium.Dev Biol. 2002 Aug 1;248(1):1-12. doi: 10.1006/dbio.2002.0715. Dev Biol. 2002. PMID: 12142016
-
Adenylyl cyclase expression and regulation during the differentiation of Dictyostelium discoideum.IUBMB Life. 2004 Sep;56(9):541-6. doi: 10.1080/15216540400013887. IUBMB Life. 2004. PMID: 15590560 Review.
Cited by
-
Evolutionary crossroads in developmental biology: Dictyostelium discoideum.Development. 2011 Feb;138(3):387-96. doi: 10.1242/dev.048934. Development. 2011. PMID: 21205784 Free PMC article. Review.
-
The hybrid type polyketide synthase SteelyA is required for cAMP signalling in early Dictyostelium development.PLoS One. 2014 Sep 15;9(9):e106634. doi: 10.1371/journal.pone.0106634. eCollection 2014. PLoS One. 2014. PMID: 25222736 Free PMC article.
-
The Evolution of Aggregative Multicellularity and Cell-Cell Communication in the Dictyostelia.J Mol Biol. 2015 Nov 20;427(23):3722-33. doi: 10.1016/j.jmb.2015.08.008. Epub 2015 Aug 15. J Mol Biol. 2015. PMID: 26284972 Free PMC article. Review.
-
Naegleria genus pangenome reveals new structural and functional insights into the versatility of these free-living amoebae.Front Microbiol. 2023 Feb 1;13:1056418. doi: 10.3389/fmicb.2022.1056418. eCollection 2022. Front Microbiol. 2023. PMID: 36817109 Free PMC article.
-
Evolution of developmental cyclic adenosine monophosphate signaling in the Dictyostelia from an amoebozoan stress response.Dev Growth Differ. 2011 May;53(4):452-62. doi: 10.1111/j.1440-169X.2011.01263.x. Dev Growth Differ. 2011. PMID: 21585352 Free PMC article. Review.
References
-
- Linder J. U., Schultz J. E. (2003) Cellular Signalling 15, 1081–1089 - PubMed
-
- Baker D. A., Kelly J. M. (2004) Trends Parasitol. 20, 227–232 - PubMed
-
- Chen Y., Cann M. J., Litvin T. N., Iourgenko V., Sinclair M. L., Levin L. R., Buck J. (2000) Science 289, 625–628 - PubMed
-
- Kim H. J., Chang W. T., Meima M., Gross J. D., Schaap P. (1998) J. Biol. Chem. 273, 30859–30862 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

