Structural basis for hormone recognition by the Human CRFR2{alpha} G protein-coupled receptor
- PMID: 20966082
- PMCID: PMC3001015
- DOI: 10.1074/jbc.M110.186072
Structural basis for hormone recognition by the Human CRFR2{alpha} G protein-coupled receptor
Abstract
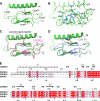
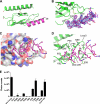
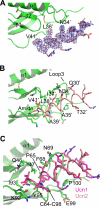
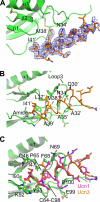
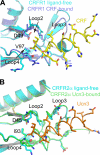
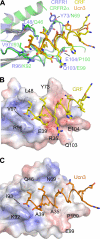
The mammalian corticotropin releasing factor (CRF)/urocortin (Ucn) peptide hormones include four structurally similar peptides, CRF, Ucn1, Ucn2, and Ucn3, that regulate stress responses, metabolism, and cardiovascular function by activating either of two related class B G protein-coupled receptors, CRFR1 and CRFR2. CRF and Ucn1 activate both receptors, whereas Ucn2 and Ucn3 are CRFR2-selective. The molecular basis for selectivity is unclear. Here, we show that the purified N-terminal extracellular domains (ECDs) of human CRFR1 and the CRFR2α isoform are sufficient to discriminate the peptides, and we present three crystal structures of the CRFR2α ECD bound to each of the Ucn peptides. The CRFR2α ECD forms the same fold observed for the CRFR1 and mouse CRFR2β ECDs but contains a unique N-terminal α-helix formed by its pseudo signal peptide. The CRFR2α ECD peptide-binding site architecture is similar to that of CRFR1, and binding of the α-helical Ucn peptides closely resembles CRF binding to CRFR1. Comparing the electrostatic surface potentials of the ECDs suggests a charge compatibility mechanism for ligand discrimination involving a single amino acid difference in the receptors (CRFR1 Glu104/CRFR2α Pro-100) at a site proximate to peptide residue 35 (Arg in CRF/Ucn1, Ala in Ucn2/3). CRFR1 Glu-104 acts as a selectivity filter preventing Ucn2/3 binding because the nonpolar Ala-35 is incompatible with the negatively charged Glu-104. The structures explain the mechanisms of ligand recognition and discrimination and provide a molecular template for the rational design of therapeutic agents selectively targeting these receptors.
Figures

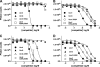
Similar articles
-
Cardiac CRFR1 Expression Is Elevated in Human Heart Failure and Modulated by Genetic Variation and Alternative Splicing.Endocrinology. 2016 Dec;157(12):4865-4874. doi: 10.1210/en.2016-1448. Epub 2016 Oct 18. Endocrinology. 2016. PMID: 27754786 Free PMC article.
-
Molecular recognition of corticotropin-releasing factor by its G-protein-coupled receptor CRFR1.J Biol Chem. 2008 Nov 21;283(47):32900-12. doi: 10.1074/jbc.M805749200. Epub 2008 Sep 17. J Biol Chem. 2008. PMID: 18801728 Free PMC article.
-
Glucocorticoids differentially regulate the expression of CRFR1 and CRFR2α in MIN6 insulinoma cells and rodent islets.Endocrinology. 2011 Jan;152(1):138-50. doi: 10.1210/en.2010-0791. Epub 2010 Nov 24. Endocrinology. 2011. PMID: 21106875 Free PMC article.
-
Urocortins: CRF's siblings and their potential role in anxiety, depression and alcohol drinking behavior.Alcohol. 2012 Jun;46(4):349-57. doi: 10.1016/j.alcohol.2011.10.007. Epub 2012 Mar 22. Alcohol. 2012. PMID: 22444954 Free PMC article. Review.
-
The CRF/Urocortin systems as therapeutic targets for alcohol use disorders.Int Rev Neurobiol. 2024;178:97-152. doi: 10.1016/bs.irn.2024.08.002. Epub 2024 Aug 28. Int Rev Neurobiol. 2024. PMID: 39523064 Review.
Cited by
-
New Insights into the Structure and Function of Class B1 GPCRs.Endocr Rev. 2023 May 8;44(3):492-517. doi: 10.1210/endrev/bnac033. Endocr Rev. 2023. PMID: 36546772 Free PMC article.
-
Crystal structure of the PAC1R extracellular domain unifies a consensus fold for hormone recognition by class B G-protein coupled receptors.PLoS One. 2011;6(5):e19682. doi: 10.1371/journal.pone.0019682. Epub 2011 May 19. PLoS One. 2011. PMID: 21625560 Free PMC article.
-
Selective CGRP and adrenomedullin peptide binding by tethered RAMP-calcitonin receptor-like receptor extracellular domain fusion proteins.Protein Sci. 2013 Dec;22(12):1775-85. doi: 10.1002/pro.2377. Epub 2013 Oct 19. Protein Sci. 2013. PMID: 24115156 Free PMC article.
-
Calcitonin and Amylin Receptor Peptide Interaction Mechanisms: INSIGHTS INTO PEPTIDE-BINDING MODES AND ALLOSTERIC MODULATION OF THE CALCITONIN RECEPTOR BY RECEPTOR ACTIVITY-MODIFYING PROTEINS.J Biol Chem. 2016 Apr 15;291(16):8686-700. doi: 10.1074/jbc.M115.713628. Epub 2016 Feb 19. J Biol Chem. 2016. PMID: 26895962 Free PMC article.
-
Structural and Functional Insights into CRF Peptides and Their Receptors.Biology (Basel). 2024 Feb 13;13(2):120. doi: 10.3390/biology13020120. Biology (Basel). 2024. PMID: 38392338 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

