SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse
- PMID: 20966168
- PMCID: PMC3031475
- DOI: 10.1182/blood-2010-03-273011
SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse
Abstract
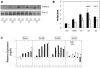
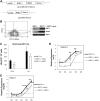
SIRT1 is a founding member of a sirtuin family of 7 proteins and histone deacetylases. It is involved in cellular resistance to stress, metabolism, differentiation, aging, and tumor suppression. SIRT1(-/-) mice demonstrate embryonic and postnatal development defects. We examined hematopoietic and endothelial cell differentiation of SIRT1(-/-) mouse embryonic stem cells (ESCs) in vitro, and hematopoietic progenitors in SIRT1(+/+)(+/-), and (-/-) mice. SIRT1(-/-) ESCs formed fewer mature blast cell colonies. Replated SIRT1(-/-) blast colony-forming cells demonstrated defective hematopoietic potential. Endothelial cell production was unaltered, but there were defects in formation of a primitive vascular network from SIRT1(-/-)-derived embryoid bodies. Development of primitive and definitive progenitors derived from SIRT1(-/-) ESCs were also delayed and/or defective. Differentiation delay/defects were associated with delayed capacity to switch off Oct4, Nanog and Fgf5 expression, decreased β-H1 globin, β-major globin, and Scl gene expression, and reduced activation of Erk1/2. Ectopic expression of SIRT1 rescued SIRT1(-/-) ESC differentiation deficiencies. SIRT1(-/-) yolk sacs manifested fewer primitive erythroid precursors. SIRT1(-/-) and SIRT1(+/-) adult marrow had decreased numbers and cycling of hematopoietic progenitors, effects more apparent at 5%, than at 20%, oxygen tension, and these progenitors survived less well in vitro under conditions of delayed growth factor addition. This suggests a role for SIRT1 in ESC differentiation and mouse hematopoiesis.
Figures





References
-
- Smith AG, Heath JK, Donaldson DD, et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336(6200):688–690. - PubMed
-
- Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995;7(6):862–869. - PubMed
-
- Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G. A common precursor for hematopoietic and endothelial cells. Development. 1998;125(4):725–732. - PubMed
-
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273(2):793–798. - PubMed
-
- Guarente L, Picard F. Calorie restriction—the SIR2 connection. Cell. 2005;120(4):473–482. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

