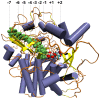
Catalytic mechanism of cellulose degradation by a cellobiohydrolase, CelS
- PMID: 20967294
- PMCID: PMC2953488
- DOI: 10.1371/journal.pone.0012947
Catalytic mechanism of cellulose degradation by a cellobiohydrolase, CelS
Abstract
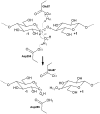
The hydrolysis of cellulose is the bottleneck in cellulosic ethanol production. The cellobiohydrolase CelS from Clostridium thermocellum catalyzes the hydrolysis of cello-oligosaccharides via inversion of the anomeric carbon. Here, to examine key features of the CelS-catalyzed reaction, QM/MM (SCCDFTB/MM) simulations are performed. The calculated free energy profile for the reaction possesses a 19 kcal/mol barrier. The results confirm the role of active site residue Glu87 as the general acid catalyst in the cleavage reaction and show that Asp255 may act as the general base. A feasible position in the reactant state of the water molecule responsible for nucleophilic attack is identified. Sugar ring distortion as the reaction progresses is quantified. The results provide a computational approach that may complement the experimental design of more efficient enzymes for biofuel production.
Conflict of interest statement
Figures
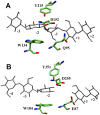
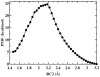
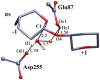
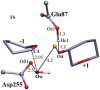
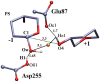
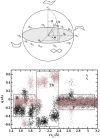
 , and q
, and q ) sampled by molecular dynamics trajectories along the reaction coordinate rc2. q
) sampled by molecular dynamics trajectories along the reaction coordinate rc2. q and q
and q values are shown in gray and black, respectively. Regions corresponding to reactant (R), around transition state (TS), and product (R) are highlighted by boxes.
values are shown in gray and black, respectively. Regions corresponding to reactant (R), around transition state (TS), and product (R) are highlighted by boxes.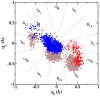
 C
C conformation.
conformation.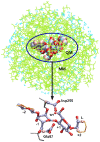
References
-
- Sticklen M. Plant genetic engineering for biofuel production: towards affordable cellulosic ethanol. Nat Rev Gent. 2008;9:433–443. - PubMed
-
- Himmel M, Ding S, Johnson D, Adney W, Nimlos M, et al. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science. 2007;315:804. - PubMed
-
- Beguin P, Aubert J. The biological degradation of cellulose. FEMS Microbiol Rev. 1994;13:25–58. - PubMed
-
- Creuzet N, Berenger J, Frixon C. Characterization of exoglucanase and synergistic hydrolysis of cellulose in Clostridium stercorarium. FEMS Microbiol Lett. 1983;20:347–350.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

