Constitutive phosphorylation by protein kinase C regulates D1 dopamine receptor signaling
- PMID: 20969574
- PMCID: PMC2996484
- DOI: 10.1111/j.1471-4159.2010.07074.x
Constitutive phosphorylation by protein kinase C regulates D1 dopamine receptor signaling
Abstract
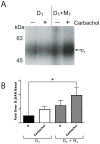
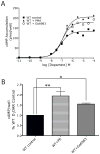
The D(1) dopamine receptor (D(1) DAR) is robustly phosphorylated by multiple protein kinases, yet the phosphorylation sites and functional consequences of these modifications are not fully understood. Here, we report that the D(1) DAR is phosphorylated by protein kinase C (PKC) in the absence of agonist stimulation. Phosphorylation of the D(1) DAR by PKC is constitutive in nature, can be induced by phorbol ester treatment or through activation of Gq-mediated signal transduction pathways, and is abolished by PKC inhibitors. We demonstrate that most, but not all, isoforms of PKC are capable of phosphorylating the receptor. To directly assess the functional role of PKC phosphorylation of the D(1) DAR, a site-directed mutagenesis approach was used to identify the PKC sites within the receptor. Five serine residues were found to mediate the PKC phosphorylation. Replacement of these residues had no effect on D(1) DAR expression or agonist-induced desensitization; however, G protein coupling and cAMP accumulation were significantly enhanced in PKC-null D(1) DAR. Thus, constitutive or heterologous PKC phosphorylation of the D(1) DAR dampens dopamine activation of the receptor, most likely occurring in a context-specific manner, mediated by the repertoire of PKC isozymes within the cell.
© 2010 The Authors. Journal of Neurochemistry © 2010 International Society for Neurochemistry.
Figures






References
-
- Bouvier M. Cross-talk between second messengers. Ann N Y Acad Sci. 1990;594:120–129. - PubMed
-
- Chen CH, Paing MM, Trejo J. Termination of protease-activated receptor-1 signaling by beta-arrestins is independent of receptor phosphorylation. J Biol Chem. 2004;279:10020–10031. - PubMed
-
- Ferguson SS. Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol Rev. 2001;53:1–24. - PubMed
-
- Fredriksson R, Schioth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67:1414–1425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

