Transcriptional control of invariant NKT cell development
- PMID: 20969594
- PMCID: PMC2965566
- DOI: 10.1111/j.1600-065X.2010.00962.x
Transcriptional control of invariant NKT cell development
Abstract
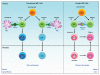
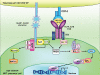
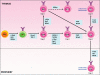
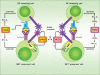
Invariant natural killer T (iNKT) cells comprise a rare lymphocyte sublineage with phenotypic and functional properties similar to T and NK cells. Akin to conventional αβ T cells, their development occurs primarily in the thymus, where they originate from CD4(+) CD8(+) double positive (DP) progenitors. However, the selection of iNKT cells is unique in that it is mediated by homotypic interactions of DP cells and recognition of glycolipid antigen-CD1d complexes. Additionally, iNKT cells acquire an activated innate-like phenotype during development that allows them to release cytokines rapidly following antigen exposure. Given their hybrid features, it is not surprising that the developmental program of iNKT cells partially overlaps with that of T and NK cells. Several recent reports have provided new and exciting insights into the developmental mechanisms that direct natural killer T (NKT) cell lineage commitment and maturation. In this review, we provide a discussion of the NKT cell developmental program with an emphasis on the signaling mechanisms and transcription factors that influence the ontogeny of this lineage. Continued investigations into the complex interplay of these transcription factors and their relationship with other extracellular and intracellular signaling molecules will undoubtedly provide important clues into the biology of this unusual T-cell lineage.
© 2010 John Wiley & Sons A/S.
Figures

References
-
- Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 2010;11:197–206. - PubMed
-
- Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7:505–518. - PubMed
-
- Kawano T, et al. CD1d-restricted and TCR-mediated activation of Valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. - PubMed
-
- Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what’s in a name. Nat Rev Immunol. 2004;4:231–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

