Regulation of the Ifng locus in the context of T-lineage specification and plasticity
- PMID: 20969595
- PMCID: PMC3096439
- DOI: 10.1111/j.1600-065X.2010.00961.x
Regulation of the Ifng locus in the context of T-lineage specification and plasticity
Abstract
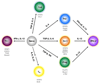
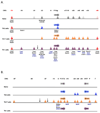
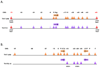
Study of the development of distinct CD4(+) T-cell subsets from naive precursors continues to provide excellent opportunities for dissection of mechanisms that control lineage-specific gene expression or repression. Whereas it had been thought that the induction of transcription networks that control T-lineage commitment were highly stable, reinforced by epigenetic processes that confer heritability of functional phenotypes by the progeny of mature T cells, recent findings support a more dynamic view of T-lineage commitment. Here, we highlight advances in the mapping and functional characterization of cis elements in the Ifng locus that have provided new insights into the control of the chromatin structure and transcriptional activity of this signature T-helper 1 cell gene. We also examine epigenetic features of the Ifng locus that have evolved to enable its reprogramming for expression by other T-cell subsets, particularly T-helper 17 cells, and contrast features of the Ifng locus with those of the Il17a-Il17f locus, which appears less promiscuous.
© 2010 John Wiley & Sons A/S.
Figures
References
-
- Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. - PubMed
-
- Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. - PubMed
-
- Weaver CT, Murphy KM. T-cell subsets: the more the merrier. Curr Biol. 2007;17:R61–R63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

