A multi-level study of recombinant Pichia pastoris in different oxygen conditions
- PMID: 20969759
- PMCID: PMC2987880
- DOI: 10.1186/1752-0509-4-141
A multi-level study of recombinant Pichia pastoris in different oxygen conditions
Abstract
Background: Yeasts are attractive expression platforms for many recombinant proteins, and there is evidence for an important interrelation between the protein secretion machinery and environmental stresses. While adaptive responses to such stresses are extensively studied in Saccharomyces cerevisiae, little is known about their impact on the physiology of Pichia pastoris. We have recently reported a beneficial effect of hypoxia on recombinant Fab secretion in P. pastoris chemostat cultivations. As a consequence, a systems biology approach was used to comprehensively identify cellular adaptations to low oxygen availability and the additional burden of protein production. Gene expression profiling was combined with proteomic analyses and the 13C isotope labelling based experimental determination of metabolic fluxes in the central carbon metabolism.
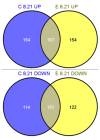
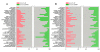
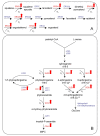
Results: The physiological adaptation of P. pastoris to hypoxia showed distinct traits in relation to the model yeast S. cerevisiae. There was a positive correlation between the transcriptomic, proteomic and metabolic fluxes adaptation of P. pastoris core metabolism to hypoxia, yielding clear evidence of a strong transcriptional regulation component of key pathways such as glycolysis, pentose phosphate pathway and TCA cycle. In addition, the adaptation to reduced oxygen revealed important changes in lipid metabolism, stress responses, as well as protein folding and trafficking.
Conclusions: This systems level study helped to understand the physiological adaptations of cellular mechanisms to low oxygen availability in a recombinant P. pastoris strain. Remarkably, the integration of data from three different levels allowed for the identification of differences in the regulation of the core metabolism between P. pastoris and S. cerevisiae. Detailed comparative analysis of the transcriptomic data also led to new insights into the gene expression profiles of several cellular processes that are not only susceptible to low oxygen concentrations, but might also contribute to enhanced protein secretion.
Figures


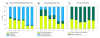
References
-
- Porro D, Mattanovich D. Recombinant protein production in yeasts. Methods Mol Biol. 2004;267:241–258. - PubMed
-
- Knijnenburg T, Daran J, van den Broek M, Daran-Lapujade P, de Winde J, Pronk J, Reinders M, Wessels L. Combinatorial effects of environmental parameters on transcriptional regulation in Saccharomyces cerevisiae: a quantitative analysis of a compendium of chemostat-based transcriptome data. BMC Genomics. 2009;10:53. doi: 10.1186/1471-2164-10-53. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

