Differential phenotyping of Brucella species using a newly developed semi-automated metabolic system
- PMID: 20969797
- PMCID: PMC2984481
- DOI: 10.1186/1471-2180-10-269
Differential phenotyping of Brucella species using a newly developed semi-automated metabolic system
Abstract
Background: A commercial biotyping system (Taxa Profile™, Merlin Diagnostika) testing the metabolization of various substrates by bacteria was used to determine if a set of phenotypic features will allow the identification of members of the genus Brucella and their differentiation into species and biovars.
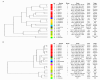
Results: A total of 191 different amines, amides, amino acids, other organic acids and heterocyclic and aromatic substrates (Taxa Profile™ A), 191 different mono-, di-, tri- and polysaccharides and sugar derivates (Taxa Profile™ C) and 95 amino peptidase- and protease-reactions, 76 glycosidase-, phosphatase- and other esterase-reactions, and 17 classic reactions (Taxa Profile™ E) were tested with the 23 reference strains representing the currently known species and biovars of Brucella and a collection of 60 field isolates. Based on specific and stable reactions a 96-well "Brucella identification and typing" plate (Micronaut™) was designed and re-tested in 113 Brucella isolates and a couple of closely related bacteria.Brucella species and biovars revealed characteristic metabolic profiles and each strain showed an individual pattern. Due to their typical metabolic profiles a differentiation of Brucella isolates to the species level could be achieved. The separation of B. canis from B. suis bv 3, however, failed. At the biovar level, B. abortus bv 4, 5, 7 and B. suis bv 1-5 could be discriminated with a specificity of 100%. B. melitensis isolates clustered in a very homogenous group and could not be resolved according to their assigned biovars.
Conclusions: The comprehensive testing of metabolic activity allows cluster analysis within the genus Brucella. The biotyping system developed for the identification of Brucella and differentiation of its species and biovars may replace or at least complement time-consuming tube testing especially in case of atypical strains. An easy to handle identification software facilitates the applicability of the Micronaut™ system for microbiology laboratories.
Figures


References
-
- Al Dahouk S, Tomaso H, Nöckler K, Neubauer H, Frangoulidis D. Laboratory-based diagnosis of brucellosis - a review of the literature. Part I: techniques for direct detection and identification of Brucella spp. Clin Lab. 2003;49:487–505. - PubMed
-
- Alton GG, Jones LM, Angus RD, Verger JM. Techniques for the brucellosis laboratory. Paris : Institut National de la Recherche Agronomique; 1988.
-
- Morgan WJB, Corbel MJ. Recommendations for the description of species and biotypes of the genus Brucella. Develop Biol Standard. 1976;31:27–37. - PubMed
-
- Corbel MJ, Brinley-Morgan WJ. In: Bergey's Manual of Systematic Bacteriology. Krieg NR, Holt JG, editor. Vol. 1. Baltimore: Williams and Wilkins; 1984. Genus Brucella; p. 370.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

