Patients on HAART often have an excess of unintegrated HIV DNA: implications for monitoring reservoirs
- PMID: 20970154
- PMCID: PMC3253773
- DOI: 10.1016/j.virol.2010.08.024
Patients on HAART often have an excess of unintegrated HIV DNA: implications for monitoring reservoirs
Abstract
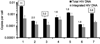
HIV establishes a latent reservoir early in infection that is resistant to anti-retroviral therapy and has a slow rate of decay. It is thought that the majority of HIV DNA in treated patients is integrated since unintegrated HIV DNA appears to be unstable. Thus, to monitor the HIV latent reservoir, total HIV DNA is commonly measured in PBMC from infected individuals. We investigated how often total approaches integrated HIV DNA in treated patients. To do this, we first assessed how accurate our integration assay is and determined the error in our measurements of total and integrated HIV DNA. We demonstrated an excess of total over integrated HIV DNA was present in a subset of patients, suggesting that measurements of total HIV DNA do not always correlate to the level of integration. Determining the cause of this excess and its frequency may have important implications for understanding HIV latent reservoir maintenance.
Copyright © 2010. Published by Elsevier Inc.
Figures




References
-
- Agosto LM, Yu JJ, Liszewski MK, Baytop C, Korokhov N, Humeau LM, O'Doherty U. The CXCR4-tropic human immunodeficiency virus envelope promotes more efficient gene delivery to resting CD4+ T cells than the vesicular stomatitis virus glycoprotein G envelope. J Virol. 2009;83(16):8153–8162. - PMC - PubMed
-
- Ansari-Lari MA, Donehower LA, Gibbs RA. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology. 1995;211(1):332–335. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

