Control of translation efficiency in yeast by codon-anticodon interactions
- PMID: 20971810
- PMCID: PMC2995412
- DOI: 10.1261/rna.2411710
Control of translation efficiency in yeast by codon-anticodon interactions
Abstract
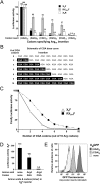
The choice of synonymous codons used to encode a polypeptide contributes to substantial differences in translation efficiency between genes. However, both the magnitude and the mechanisms of codon-mediated effects are unknown, as neither the effects of individual codons nor the parameters that modulate codon-mediated regulation are understood, particularly in eukaryotes. To explore this problem in Saccharomyces cerevisiae, we performed the first systematic analysis of codon effects on expression. We find that the arginine codon CGA is strongly inhibitory, resulting in progressively and sharply reduced expression with increased CGA codon dosage. CGA-mediated inhibition of expression is primarily due to wobble decoding of CGA, since it is nearly completely suppressed by coexpression of an exact match anticodon-mutated tRNA(Arg(UCG)), and is associated with generation of a smaller RNA fragment, likely due to endonucleolytic cleavage at a stalled ribosome. Moreover, CGA codon pairs are more effective inhibitors of expression than individual CGA codons. These results directly implicate decoding by the ribosome and interactions at neighboring sites within the ribosome as mediators of codon-specific translation efficiency.
Figures




References
-
- Alexandrov A, Vignali M, LaCount DJ, Quartley E, de Vries C, De Rosa D, Babulski J, Mitchell SF, Schoenfeld LW, Fields S, et al. 2004. A facile method for high-throughput co-expression of protein pairs. Mol Cell Proteomics 3: 934–938 - PubMed
-
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96 - PubMed
-
- Bennetzen JL, Hall BD 1982. Codon selection in yeast. J Biol Chem 257: 3026–3031 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
