Persistence of seed-based activity following segmentation of a microRNA guide strand
- PMID: 20971811
- PMCID: PMC2995395
- DOI: 10.1261/rna.2296210
Persistence of seed-based activity following segmentation of a microRNA guide strand
Abstract
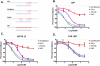
microRNAs are ∼ 22 nucleotide regulatory RNAs that are processed into duplexes from hairpin structures and incorporated into Argonaute proteins. Here, we show that a nick in the middle of the guide strand of an miRNA sequence allows for seed-based targeting characteristic of miRNA activity. Insertion of an inverted abasic, a dye, or a small gap between the two segments still permits target knockdown. While activity from the seed region of the segmented miRNA is apparent, activity from the 3' half of the guide strand is impaired, suggesting that an intact guide backbone is required for contribution from the 3' half. miRNA activity was also observed following nicking of a miRNA precursor. These results illustrate a structural flexibility in miRNA duplexes and may have applications in the design of miRNA mimetics.
Figures



References
-
- Diederichs S, Haber DA 2007. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131: 1097–1108 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
