A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif
- PMID: 20971849
- PMCID: PMC3003379
- DOI: 10.1074/jbc.M110.173161
A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif
Abstract
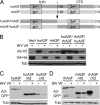
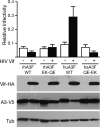
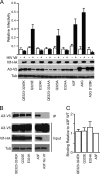
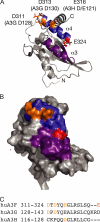
Human APOBEC3F (huA3F) potently restricts the infectivity of HIV-1 in the absence of the viral accessory protein virion infectivity factor (Vif). Vif functions to preserve viral infectivity by triggering the degradation of huA3F but not rhesus macaque A3F (rhA3F). Here, we use a combination of deletions, chimeras, and systematic mutagenesis between huA3F and rhA3F to identify Glu(324) as a critical determinant of huA3F susceptibility to HIV-1 Vif-mediated degradation. A structural model of the C-terminal deaminase domain of huA3F indicates that Glu(324) is a surface residue within the α4 helix adjacent to residues corresponding to other known Vif susceptibility determinants in APOBEC3G and APOBEC3H. This structural clustering suggests that Vif may bind a conserved surface present in multiple APOBEC3 proteins.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

