Genetic improvement of Bacillus licheniformis strains for efficient deproteinization of shrimp shells and production of high-molecular-mass chitin and chitosan
- PMID: 20971870
- PMCID: PMC3008253
- DOI: 10.1128/AEM.01404-10
Genetic improvement of Bacillus licheniformis strains for efficient deproteinization of shrimp shells and production of high-molecular-mass chitin and chitosan
Abstract
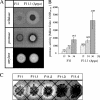
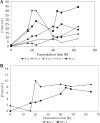
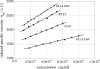
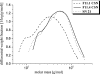
By targeted deletion of the polyglutamate operon (pga) in Bacillus licheniformis F11, a derivative form, F11.1 (Δpga), was obtained that, along with lacking polyglutamate (PGA) formation, displayed enhanced proteolytic activities. The phenotypic properties were maintained in a strain in which the chiBA operon was additionally deleted: F11.4 (ΔchiBA Δpga). These genetically modified strains, carrying the Δpga deletion either alone (F11.1) or together with the ΔchiBA (F11.4) deletion, were used in fermentations (20-liter scale) aiming at the deproteinization of shrimp shells in order to obtain long-chain chitin. After chemical deacetylation, the resulting chitosan samples were analyzed by nuclear magnetic resonance spectroscopy, size exclusion chromatography, and viscometry and compared to a chitosan preparation that was produced in parallel by chemical methods by a commercial chitosan supplier (GSRmbH). Though faint lipid impurities were present in the fermented polysaccharides, the viscosity of the material produced with the double-deletion mutant F11.4 (Δpga ΔchiBA) was higher than that of the chemically produced and commercially available samples (Cognis GmbH). Thus, enhanced proteolytic activities and a lack of chitinase activity render the double mutant F11.4 a powerful tool for the production of long-chain chitosan.
Figures



References
-
- Bautista, J., M. Jover, J. F. Gutierrez, R. Corpas, O. Cremades, E. Fontiveros, F. Iglesias, and J. Vega. 2001. Preparation of crayfish chitin by in situ lactic acid production. Process Biochem. 37:229-234.
-
- Bhaskar, N., P. V. Suresh, P. Z. Sakhare, and N. M. Sachindra. 2007. Shrimp biowaste fermentation with Pediococcus acidolactici CFR2182: optimization of fermentation conditions by response surface methodology and effect of optimized conditions on deproteination/demineralization and carotenoid recovery. Enzyme Microbiol. Technol. 40:1427-1434.
-
- Brugnerotto, J., J. Desbrières, L. Heux, K. Mazeau, and M. Rinaudo. 2001. Overview on structural characterization of chitosan molecules in relation with their behavior in solution. Macromol. Symp. 168:1-20.
-
- Chang, S., and S. N. Cohen. 1979. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol. Gen. Genet. 168:111-115. - PubMed
-
- Chen, H. C., K. A. Phang, S. D. Wu, and W. J. Mau. 2001. Isolation of chitin from shrimp shells deproteinized by Candida parapsilosis CCRC 20515. Food Sci. Agric. Chem. 3:114-120.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

