Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain
- PMID: 20971871
- PMCID: PMC3008239
- DOI: 10.1128/AEM.01233-10
Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain
Abstract
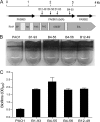
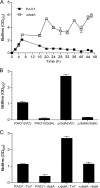
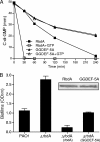
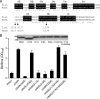
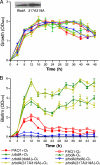
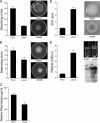
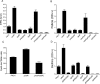
Pseudomonas aeruginosa encodes many enzymes that are potentially associated with the synthesis or degradation of the widely conserved second messenger cyclic-di-GMP (c-di-GMP). In this study, we show that mutation of rbdA, which encodes a fusion protein consisting of PAS-PAC-GGDEF-EAL multidomains, results in decreased biofilm dispersal. RbdA contains a highly conserved GGDEF domain and EAL domain, which are involved in the synthesis and degradation of c-di-GMP, respectively. However, in vivo and in vitro analyses show that the full-length RbdA protein only displays phosphodiesterase activity, causing c-di-GMP degradation. Further analysis reveals that the GGDEF domain of RbdA plays a role in activating the phosphodiesterase activity of the EAL domain in the presence of GTP. Moreover, we show that deletion of the PAS domain or substitution of the key residues implicated in sensing low-oxygen stress abrogates the functionality of RbdA. Subsequent study showed that RbdA is involved in positive regulation of bacterial motility and production of rhamnolipids, which are associated with biofilm dispersal, and in negative regulation of production of exopolysaccharides, which are required for biofilm formation. These data indicate that the c-di-GMP-degrading regulatory protein RbdA promotes biofilm dispersal through its two-pronged effects on biofilm development, i.e., downregulating biofilm formation and upregulating production of the factors associated with biofilm dispersal.
Figures

References
-
- Alm, R. A., J. P. Hallinan, A. A. Watson, and J. S. Mattick. 1996. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol. Microbiol. 22:161-173. - PubMed
-
- Applegate, D. H., and J. D. Bryers. 1991. Effects of carbon and oxygen limitations and calcium concentrations on biofilm removal processes. Biotechnol. Bioeng. 37:17-25. - PubMed
-
- Boles, B. R., M. Thoendel, and P. K. Singh. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 57:1210-1223. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

