Mapping of the Neisseria meningitidis NadA cell-binding site: relevance of predicted {alpha}-helices in the NH2-terminal and dimeric coiled-coil regions
- PMID: 20971901
- PMCID: PMC3019930
- DOI: 10.1128/JB.00430-10
Mapping of the Neisseria meningitidis NadA cell-binding site: relevance of predicted {alpha}-helices in the NH2-terminal and dimeric coiled-coil regions
Abstract
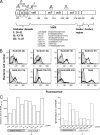
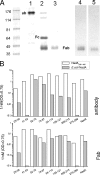
NadA is a trimeric autotransporter protein of Neisseria meningitidis belonging to the group of oligomeric coiled-coil adhesins. It is implicated in the colonization of the human upper respiratory tract by hypervirulent serogroup B N. meningitidis strains and is part of a multiantigen anti-serogroup B vaccine. Structure prediction indicates that NadA is made by a COOH-terminal membrane anchor (also necessary for autotranslocation to the bacterial surface), an intermediate elongated coiled-coil-rich stalk, and an NH(2)-terminal region involved in cell interaction. Electron microscopy analysis and structure prediction suggest that the apical region of NadA forms a compact and globular domain. Deletion studies proved that the NH(2)-terminal sequence (residues 24 to 87) is necessary for cell adhesion. In this study, to better define the NadA cell binding site, we exploited (i) a panel of NadA mutants lacking sequences along the coiled-coil stalk and (ii) several oligoclonal rabbit antibodies, and their relative Fab fragments, directed to linear epitopes distributed along the NadA ectodomain. We identified two critical regions for the NadA-cell receptor interaction with Chang cells: the NH(2) globular head domain and the NH(2) dimeric intrachain coiled-coil α-helices stemming from the stalk. This raises the importance of different modules within the predicted NadA structure. The identification of linear epitopes involved in receptor binding that are able to induce interfering antibodies reinforces the importance of NadA as a vaccine antigen.
Figures


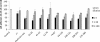
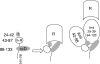
Similar articles
-
Structure of the meningococcal vaccine antigen NadA and epitope mapping of a bactericidal antibody.Proc Natl Acad Sci U S A. 2014 Dec 2;111(48):17128-33. doi: 10.1073/pnas.1419686111. Epub 2014 Nov 17. Proc Natl Acad Sci U S A. 2014. PMID: 25404323 Free PMC article.
-
Neisserial adhesin A (NadA) binds human Siglec-5 and Siglec-14 with high affinity and promotes bacterial adhesion/invasion.mBio. 2024 Aug 14;15(8):e0110724. doi: 10.1128/mbio.01107-24. Epub 2024 Jul 23. mBio. 2024. PMID: 39041817 Free PMC article.
-
NadA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed by Meningococcus B Vaccine-Elicited Human Antibodies.mBio. 2018 Oct 16;9(5):e01914-18. doi: 10.1128/mBio.01914-18. mBio. 2018. PMID: 30327444 Free PMC article.
-
Neisseria meningitidis adhesin NadA targets beta1 integrins: functional similarity to Yersinia invasin.J Biol Chem. 2011 Jun 10;286(23):20536-46. doi: 10.1074/jbc.M110.188326. Epub 2011 Apr 6. J Biol Chem. 2011. PMID: 21471204 Free PMC article.
-
Structure and biology of trimeric autotransporter adhesins.Adv Exp Med Biol. 2011;715:143-58. doi: 10.1007/978-94-007-0940-9_9. Adv Exp Med Biol. 2011. PMID: 21557062 Review.
Cited by
-
Rapid profiling of the antigen regions recognized by serum antibodies using massively parallel sequencing of antigen-specific libraries.PLoS One. 2014 Dec 4;9(12):e114159. doi: 10.1371/journal.pone.0114159. eCollection 2014. PLoS One. 2014. PMID: 25473968 Free PMC article.
-
Type V Secretion Systems: An Overview of Passenger Domain Functions.Front Microbiol. 2019 May 31;10:1163. doi: 10.3389/fmicb.2019.01163. eCollection 2019. Front Microbiol. 2019. PMID: 31214135 Free PMC article. Review.
-
Biological Functions of the Secretome of Neisseria meningitidis.Front Cell Infect Microbiol. 2017 Jun 16;7:256. doi: 10.3389/fcimb.2017.00256. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28670572 Free PMC article. Review.
-
Type 5 secretion system antigens as vaccines against Gram-negative bacterial infections.NPJ Vaccines. 2024 Sep 1;9(1):159. doi: 10.1038/s41541-024-00953-6. NPJ Vaccines. 2024. PMID: 39218947 Free PMC article. Review.
-
Human protective response induced by meningococcus B vaccine is mediated by the synergy of multiple bactericidal epitopes.Sci Rep. 2018 Feb 27;8(1):3700. doi: 10.1038/s41598-018-22057-7. Sci Rep. 2018. PMID: 29487324 Free PMC article.
References
-
- Brooks, M. J., J. L. Sedillo, N. Wagner, W. Wang, A. S. Attia, H. Wong, C. A. Laurence, E. J. Hansen, and S. D. Gray-Owen. 2008. Moraxella catarrhalis binding to host cellular receptors is mediated by sequence-specific determinants not conserved among all UspA1 protein variants. Infect. Immun. 76:5322-5329. - PMC - PubMed
-
- Capecchi, B., J. Adu-Bobie, F. Di Marcello, L. Ciucchi, V. Masignani, A. Taddei, R. Rappuoli, M. Pizza, and B. Arico. 2005. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol. Microbiol. 55:687-698. - PubMed
-
- Comanducci, M., S. Bambini, B. Brunelli, J. Adu-Bobie, B. Aricò, B. Capecchi, M. M. Giuliani, V. Masignani, L. Santini, S. Savino, D. M. Granoff, D. A. Caugant, M. Pizza, R. Rappuoli, and M. Mora. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 195:1445-1454. - PMC - PubMed
-
- Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13:199-205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

