The Rcs signal transduction pathway is triggered by enterobacterial common antigen structure alterations in Serratia marcescens
- PMID: 20971912
- PMCID: PMC3019949
- DOI: 10.1128/JB.00839-10
The Rcs signal transduction pathway is triggered by enterobacterial common antigen structure alterations in Serratia marcescens
Abstract
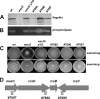
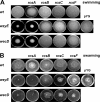
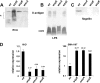
The enterobacterial common antigen (ECA) is a highly conserved exopolysaccharide in Gram-negative bacteria whose role remains largely uncharacterized. In a previous work, we have demonstrated that disrupting the integrity of the ECA biosynthetic pathway imposed severe deficiencies to the Serratia marcescens motile (swimming and swarming) capacity. In this work, we show that alterations in the ECA structure activate the Rcs phosphorelay, which results in the repression of the flagellar biogenesis regulatory cascade. In addition, a detailed analysis of wec cluster mutant strains, which provoke the disruption of the ECA biosynthesis at different levels of the pathway, suggests that the absence of the periplasmic ECA cyclic structure could constitute a potential signal detected by the RcsF-RcsCDB phosphorelay. We also identify SMA1167 as a member of the S. marcescens Rcs regulon and show that high osmolarity induces Rcs activity in this bacterium. These results provide a new perspective from which to understand the phylogenetic conservation of ECA among enterobacteria and the basis for the virulence attenuation detected in wec mutant strains in other pathogenic bacteria.
Figures

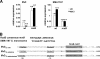


References
-
- Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42.
-
- Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques 26:824-826, 828. - PubMed
-
- Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079-1093. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

