GAPDH mediates nitrosylation of nuclear proteins
- PMID: 20972425
- PMCID: PMC2972384
- DOI: 10.1038/ncb2114
GAPDH mediates nitrosylation of nuclear proteins
Abstract
S-nitrosylation of proteins by nitric oxide is a major mode of signalling in cells. S-nitrosylation can mediate the regulation of a range of proteins, including prominent nuclear proteins, such as HDAC2 (ref. 2) and PARP1 (ref. 3). The high reactivity of the nitric oxide group with protein thiols, but the selective nature of nitrosylation within the cell, implies the existence of targeting mechanisms. Specificity of nitric oxide signalling is often achieved by the binding of nitric oxide synthase (NOS) to target proteins, either directly or through scaffolding proteins such as PSD-95 (ref. 5) and CAPON. As the three principal isoforms of NOS--neuronal NOS (nNOS), endothelial NOS (eNOS) and inducible NOS (iNOS)--are primarily non-nuclear, the mechanisms by which nuclear proteins are selectively nitrosylated have been elusive. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is physiologically nitrosylated at its Cys 150 residue. Nitrosylated GAPDH (SNO-GAPDH) binds to Siah1, which possesses a nuclear localization signal, and is transported to the nucleus. Here, we show that SNO-GAPDH physiologically transnitrosylates nuclear proteins, including the deacetylating enzyme sirtuin-1 (SIRT1), histone deacetylase-2 (HDAC2) and DNA-activated protein kinase (DNA-PK). Our findings reveal a novel mechanism for targeted nitrosylation of nuclear proteins and suggest that protein-protein transfer of nitric oxide groups may be a general mechanism in cellular signal transduction.
Figures
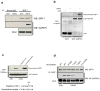


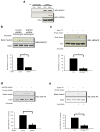
Comment in
-
Nascent nitrosylases.Nat Cell Biol. 2010 Nov;12(11):1024-6. doi: 10.1038/ncb1110-1024. Epub 2010 Oct 24. Nat Cell Biol. 2010. PMID: 20972426
References
-
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nature Rev Mol Cell Biol. 2005;6:150–166. - PubMed
-
- Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. - PubMed
-
- Yu Z, Kuncewicz T, Dubinsky WP, Kone BC. Nitric oxide-dependent negative feedback of PARP-1 trans-activation of the inducible nitric-oxide synthase gene. J Biol Chem. 2006;281:9101–9109. - PubMed
-
- Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science. 2005;310:1966–1970. - PubMed
-
- Lipton SA, et al. Cysteine regulation of protein function as exemplified by NMDA-receptor modulation. Trends Neurosci. 2002;25:474–480. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

