The TLX1 oncogene drives aneuploidy in T cell transformation
- PMID: 20972433
- PMCID: PMC2974790
- DOI: 10.1038/nm.2246
The TLX1 oncogene drives aneuploidy in T cell transformation
Abstract
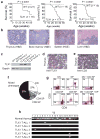
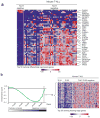
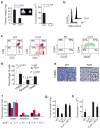
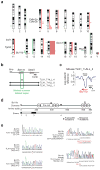
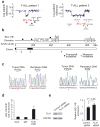
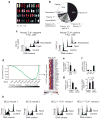
The TLX1 oncogene (encoding the transcription factor T cell leukemia homeobox protein-1) has a major role in the pathogenesis of T cell acute lymphoblastic leukemia (T-ALL). However, the specific mechanisms of T cell transformation downstream of TLX1 remain to be elucidated. Here we show that transgenic expression of human TLX1 in mice induces T-ALL with frequent deletions and mutations in Bcl11b (encoding B cell leukemia/lymphoma-11B) and identify the presence of recurrent mutations and deletions in BCL11B in 16% of human T-ALLs. Most notably, mouse TLX1 tumors were typically aneuploid and showed a marked defect in the activation of the mitotic checkpoint. Mechanistically, TLX1 directly downregulates the expression of CHEK1 (encoding CHK1 checkpoint homolog) and additional mitotic control genes and induces loss of the mitotic checkpoint in nontransformed preleukemic thymocytes. These results identify a previously unrecognized mechanism contributing to chromosomal missegregation and aneuploidy active at the earliest stages of tumor development in the pathogenesis of cancer.
Figures
Comment in
-
The T-ALL paradox in cancer.Nat Med. 2010 Nov;16(11):1185-6. doi: 10.1038/nm1110-1185. Nat Med. 2010. PMID: 21052064 No abstract available.
References
-
- Ferrando AA, et al. Prognostic importance of TLX1 (HOX11) oncogene expression in adults with T-cell acute lymphoblastic leukaemia. Lancet. 2004;363:535–536. - PubMed
-
- Ferrando AA, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. - PubMed
-
- Kees UR, et al. Expression of HOX11 in childhood T-lineage acute lymphoblastic leukaemia can occur in the absence of cytogenetic aberration at 10q24: a study from the Children's Cancer Group (CCG) Leukemia. 2003;17:887–893. - PubMed
-
- Berger R, et al. t(5;14)/HOX11L2-positive T-cell acute lymphoblastic leukemia. A collaborative study of the Groupe Francais de Cytogenetique Hematologique (GFCH) Leukemia. 2003;17:1851–1857. - PubMed
-
- Bernard OA, et al. A new recurrent and specific cryptic translocation, t(5;14)(q35;q32), is associated with expression of the Hox11L2 gene in T acute lymphoblastic leukemia. Leukemia. 2001;15 :1495–1504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

