Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes
- PMID: 20972443
- PMCID: PMC3024583
- DOI: 10.1038/nsmb.1938
Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes
Abstract
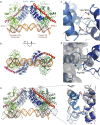
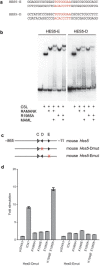
Ligand-induced proteolysis of Notch produces an intracellular effector domain that transduces essential signals by regulating the transcription of target genes. This function relies on the formation of transcriptional activation complexes that include intracellular Notch, a Mastermind co-activator and the transcription factor CSL bound to cognate DNA. These complexes form higher-order assemblies on paired, head-to-head CSL recognition sites. Here we report the X-ray structure of a dimeric human Notch1 transcription complex loaded on the paired site from the human HES1 promoter. The small interface between the Notch ankyrin domains could accommodate DNA bending and untwisting to allow a range of spacer lengths between the two sites. Cooperative dimerization occurred on the human and mouse Hes5 promoters at a sequence that diverged from the CSL-binding consensus at one of the sites. These studies reveal how promoter organizational features control cooperativity and, thus, the responsiveness of different promoters to Notch signaling.
Figures



References
-
- Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet. 2003;12(Spec No 1):R9–13. - PubMed
-
- Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. - PubMed
-
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–8. - PubMed
-
- Mumm JS, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. - PubMed
-
- Brou C, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–16. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

