Reverse phase protein microarrays advance to use in clinical trials
- PMID: 20974554
- PMCID: PMC2981612
- DOI: 10.1016/j.molonc.2010.09.003
Reverse phase protein microarrays advance to use in clinical trials
Abstract
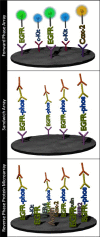
Individualizing cancer therapy for molecular targeted inhibitors requires a new class of molecular profiling technology that can map the functional state of the cancer cell signal pathways containing the drug targets. Reverse phase protein microarrays (RPMA) are a technology platform designed for quantitative, multiplexed analysis of specific phosphorylated, cleaved, or total (phosphorylated and non-phosphorylated) forms of cellular proteins from a limited amount of sample. This class of microarray can be used to interrogate tissue samples, cells, serum, or body fluids. RPMA were previously a research tool; now this technology has graduated to use in research clinical trials with clinical grade sensitivity and precision. In this review we describe the application of RPMA for multiplexed signal pathway analysis in therapeutic monitoring, biomarker discovery, and evaluation of pharmaceutical targets, and conclude with a summary of the technical aspects of RPMA construction and analysis.
Copyright © 2010 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures


Similar articles
-
Reverse-phase protein microarrays: application to biomarker discovery and translational medicine.Expert Rev Mol Diagn. 2007 Sep;7(5):625-33. doi: 10.1586/14737159.7.5.625. Expert Rev Mol Diagn. 2007. PMID: 17892368 Review.
-
Reverse phase protein microarrays and their utility in drug development.Methods Mol Biol. 2013;986:187-214. doi: 10.1007/978-1-62703-311-4_13. Methods Mol Biol. 2013. PMID: 23436414
-
Using reverse-phase protein arrays as pharmacodynamic assays for functional proteomics, biomarker discovery, and drug development in cancer.Semin Oncol. 2016 Aug;43(4):476-83. doi: 10.1053/j.seminoncol.2016.06.005. Epub 2016 Jun 15. Semin Oncol. 2016. PMID: 27663479 Free PMC article. Review.
-
Reverse phase protein arrays: mapping the path towards personalized medicine.Mol Diagn Ther. 2014 Dec;18(6):619-30. doi: 10.1007/s40291-014-0122-3. Mol Diagn Ther. 2014. PMID: 25358623 Free PMC article. Review.
-
Signaling pathway profiling by reverse-phase protein array for personalized cancer medicine.Biochim Biophys Acta. 2015 Jun;1854(6):651-7. doi: 10.1016/j.bbapap.2014.10.014. Epub 2014 Oct 27. Biochim Biophys Acta. 2015. PMID: 25448010 Review.
Cited by
-
Elevated TNFR1 and serotonin in bone metastasis are correlated with poor survival following bone metastasis diagnosis for both carcinoma and sarcoma primary tumors.Clin Cancer Res. 2013 May 1;19(9):2473-85. doi: 10.1158/1078-0432.CCR-12-3416. Epub 2013 Mar 14. Clin Cancer Res. 2013. PMID: 23493346 Free PMC article.
-
Reverse Phase Protein Arrays-Quantitative Assessment of Multiple Biomarkers in Biopsies for Clinical Use.Microarrays (Basel). 2015 Mar 24;4(2):98-114. doi: 10.3390/microarrays4020098. Microarrays (Basel). 2015. PMID: 27600215 Free PMC article. Review.
-
Phosphoprotein patterns predict trametinib responsiveness and optimal trametinib sensitisation strategies in melanoma.Cell Death Differ. 2019 Aug;26(8):1365-1378. doi: 10.1038/s41418-018-0210-8. Epub 2018 Oct 15. Cell Death Differ. 2019. PMID: 30323272 Free PMC article.
-
Reverse phase protein arrays in signaling pathways: a data integration perspective.Drug Des Devel Ther. 2015 Jul 7;9:3519-27. doi: 10.2147/DDDT.S38375. eCollection 2015. Drug Des Devel Ther. 2015. PMID: 26185419 Free PMC article. Review.
-
Systems analysis of apoptosis protein expression allows the case-specific prediction of cell death responsiveness of melanoma cells.Cell Death Differ. 2013 Nov;20(11):1521-31. doi: 10.1038/cdd.2013.106. Epub 2013 Aug 9. Cell Death Differ. 2013. PMID: 23933815 Free PMC article.
References
-
- Agarwal, R. , Gonzalez-Angulo, A.M. , Myhre, S. , Carey, M. , Lee, J.S. , Overgaard, J. , Alsner, J. , Stemke-Hale, K. , Lluch, A. , Neve, R.M. , Kuo, W.L. , Sorlie, T. , Sahin, A. , Valero, V. , Keyomarsi, K. , Gray, J.W. , Borresen-Dale, A.L. , Mills, G.B. , Hennessy, B.T. , 2009. Integrative analysis of cyclin protein levels identifies cyclin b1 as a classifier and predictor of outcomes in breast cancer. Clin. Cancer Res. 15, 3654–3662. - PMC - PubMed
-
- Alizadeh, A.A. , Eisen, M.B. , Davis, R.E. , Ma, C. , Lossos, I.S. , Rosenwald, A. , Boldrick, J.C. , Sabet, H. , Tran, T. , Yu, X. , Powell, J.I. , Yang, L. , Marti, G.E. , Moore, T. , Hudson, J. , Lu, L. , Lewis, D.B. , Tibshirani, R. , Sherlock, G. , Chan, W.C. , Greiner, T.C. , Weisenburger, D.D. , Armitage, J.O. , Warnke, R. , Staudt, L.M. , 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 403, 503–511. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

