The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells
- PMID: 20974671
- PMCID: PMC3037745
- DOI: 10.1182/blood-2010-07-298349
The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells
Abstract
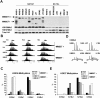
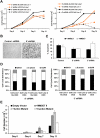
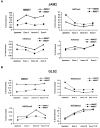
The multiple myeloma SET domain (MMSET) protein is overexpressed in multiple myeloma (MM) patients with the translocation t(4;14). Although studies have shown the involvement of MMSET/Wolf-Hirschhorn syndrome candidate 1 in development, its mode of action in the pathogenesis of MM is largely unknown. We found that MMSET is a major regulator of chromatin structure and transcription in t(4;14) MM cells. High levels of MMSET correlate with an increase in lysine 36 methylation of histone H3 and a decrease in lysine 27 methylation across the genome, leading to a more open structural state of the chromatin. Loss of MMSET expression alters adhesion properties, suppresses growth, and induces apoptosis in MM cells. Consequently, genes affected by high levels of MMSET are implicated in the p53 pathway, cell cycle regulation, and integrin signaling. Regulation of many of these genes required functional histone methyl-transferase activity of MMSET. These results implicate MMSET as a major epigenetic regulator in t(4;14)+ MM.
Figures


References
-
- Gonzalez D, van der Burg M, Garcia-Sanz R, et al. Immunoglobulin gene rearrangements and the pathogenesis of multiple myeloma. Blood. 2007;110(9):3112–3121. - PubMed
-
- Stec I, Wright TJ, van Ommen GJ, et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7(7):1071–1082. - PubMed
-
- Keats JJ, Reiman T, Belch AR, Pilarski LM. Ten years and counting: so what do we know about t(4;14)(p16;q32) multiple myeloma. Leuk Lymphoma. 2006;47(11):2289–2300. - PubMed
-
- Keats JJ, Reiman T, Maxwell CA, et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood. 2003;101(4):1520–1529. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

