Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane
- PMID: 20974814
- PMCID: PMC3003318
- DOI: 10.1083/jcb.201007098
Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane
Abstract
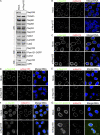
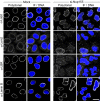
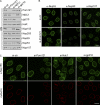
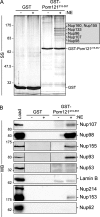
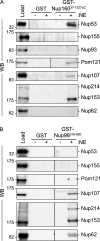
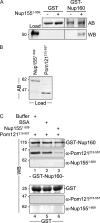
Nuclear pore complexes (NPCs) control the movement of molecules across the nuclear envelope (NE). We investigated the molecular interactions that exist at the interface between the NPC scaffold and the pore membrane. We show that key players mediating these interactions in mammalian cells are the nucleoporins Nup155 and Nup160. Nup155 depletion massively alters NE structure, causing a dramatic decrease in NPC numbers and the improper targeting of membrane proteins to the inner nuclear membrane. The role of Nup155 in assembly is likely closely linked to events at the membrane as we show that Nup155 interacts with pore membrane proteins Pom121 and NDC1. Furthermore, we demonstrate that the N terminus of Pom121 directly binds the β-propeller regions of Nup155 and Nup160. We propose a model in which the interactions of Pom121 with Nup155 and Nup160 are predicted to assist in the formation of the nuclear pore and the anchoring of the NPC to the pore membrane.
Figures



Similar articles
-
Nup53 is required for nuclear envelope and nuclear pore complex assembly.Mol Biol Cell. 2008 Apr;19(4):1753-62. doi: 10.1091/mbc.e07-08-0820. Epub 2008 Feb 6. Mol Biol Cell. 2008. PMID: 18256286 Free PMC article.
-
Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells.J Cell Biol. 2006 May 22;173(4):477-83. doi: 10.1083/jcb.200601002. Epub 2006 May 15. J Cell Biol. 2006. PMID: 16702234 Free PMC article.
-
The Nup155-mediated organisation of inner nuclear membrane proteins is independent of Nup155 anchoring to the metazoan nuclear pore complex.J Cell Sci. 2012 Sep 15;125(Pt 18):4214-8. doi: 10.1242/jcs.105809. Epub 2012 Jun 20. J Cell Sci. 2012. PMID: 22718353
-
Dunking into the Lipid Bilayer: How Direct Membrane Binding of Nucleoporins Can Contribute to Nuclear Pore Complex Structure and Assembly.Cells. 2021 Dec 20;10(12):3601. doi: 10.3390/cells10123601. Cells. 2021. PMID: 34944108 Free PMC article. Review.
-
Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes.Nucleus. 2012 May-Jun;3(3):226-36. doi: 10.4161/nucl.20148. Epub 2012 May 1. Nucleus. 2012. PMID: 22572959 Free PMC article. Review.
Cited by
-
Distinct domains in Ndc1 mediate its interaction with the Nup84 complex and the nuclear membrane.J Cell Biol. 2023 Jun 5;222(6):e202210059. doi: 10.1083/jcb.202210059. Epub 2023 May 8. J Cell Biol. 2023. PMID: 37154843 Free PMC article.
-
Improvement of Endurance Based on Muscle Fiber-Type Composition by Treatment with Dietary Apple Polyphenols in Rats.PLoS One. 2015 Jul 29;10(7):e0134303. doi: 10.1371/journal.pone.0134303. eCollection 2015. PLoS One. 2015. PMID: 26222548 Free PMC article.
-
Hepatitis C virus-induced cytoplasmic organelles use the nuclear transport machinery to establish an environment conducive to virus replication.PLoS Pathog. 2013 Oct;9(10):e1003744. doi: 10.1371/journal.ppat.1003744. Epub 2013 Oct 31. PLoS Pathog. 2013. PMID: 24204278 Free PMC article.
-
The nuclear pore complex--structure and function at a glance.J Cell Sci. 2015 Feb 1;128(3):423-9. doi: 10.1242/jcs.083246. J Cell Sci. 2015. PMID: 26046137 Free PMC article. Review.
-
Advances in the understanding of nuclear pore complexes in human diseases.J Cancer Res Clin Oncol. 2024 Jul 30;150(7):374. doi: 10.1007/s00432-024-05881-5. J Cancer Res Clin Oncol. 2024. PMID: 39080077 Free PMC article. Review.
References
-
- Abramoff M.D., Magelhaes P.J., Ram S.J.. 2004. Image processing with ImageJ. Biophotonics International. 11:36–42.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

