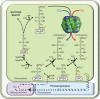
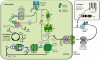
Advancing our understanding and capacity to engineer nature's CO2-sequestering enzyme, Rubisco
- PMID: 20974895
- PMCID: PMC3075749
- DOI: 10.1104/pp.110.164814
Advancing our understanding and capacity to engineer nature's CO2-sequestering enzyme, Rubisco
Figures

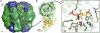

References
-
- Andersson I, Backlund A. (2008) Structure and function of Rubisco. Plant Physiol Biochem 46: 275–291 - PubMed
-
- Badger MR, Hanson D, Price GD. (2002) Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct Plant Biol 29: 161–173 - PubMed
-
- Bershtein S, Tawfik DS. (2008) Advances in laboratory evolution of enzymes. Curr Opin Chem Biol 12: 151–158 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

