Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2)
- PMID: 20974918
- PMCID: PMC2984210
- DOI: 10.1073/pnas.1008937107
Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2)
Abstract
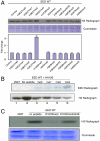
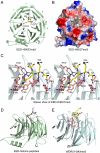
The polycomb repressive complex 2 (PRC2) is the major methyltransferase for H3K27 methylation, a modification critical for maintaining repressed gene expression programs throughout development. It has been previously shown that PRC2 maintains histone methylation patterns during DNA replication in part through its ability to bind to H3K27me3. However, the mechanism by which PRC2 recognizes H3K27me3 is unclear. Here we show that the WD40 domain of EED, a PRC2 component, is a methyllysine histone-binding domain. The crystal structures of apo-EED and EED in complex respectively with five different trimethyllysine histone peptides reveal that EED binds these peptides via the top face of its β-propeller architecture. The ammonium group of the trimethyllysine is accommodated by an aromatic cage formed by three aromatic residues, while its aliphatic chain is flanked by a fourth aromatic residue. Our structural data provide an explanation for the preferential recognition of the Ala-Arg-Lys-Ser motif-containing trimethylated H3K27, H3K9, and H1K26 marks by EED over lower methylation states and other histone methyllysine marks. More importantly, we found that binding of different histone marks by EED differentially regulates the activity and specificity of PRC2. Whereas the H3K27me3 mark stimulates the histone methyltransferase activity of PRC2, the H1K26me3 mark inhibits PRC2 methyltransferase activity on the nucleosome. Moreover, H1K26me3 binding switches the specificity of PRC2 from methylating H3K27 to EED. In addition to determining the molecular basis of EED-methyllysine recognition, our work provides the biochemical characterization of how the activity of a histone methyltransferase is oppositely regulated by two histone marks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. - PubMed
-
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. - PubMed
-
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19:266–272. - PubMed
-
- Pirrotta V, Gross DS. Epigenetic silencing mechanisms in budding yeast and fruit fly: Different paths, same destinations. Mol Cell. 2005;18:395–398. - PubMed
-
- Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

