Xenopus furry contributes to release of microRNA gene silencing
- PMID: 20974966
- PMCID: PMC2984173
- DOI: 10.1073/pnas.1008954107
Xenopus furry contributes to release of microRNA gene silencing
Abstract
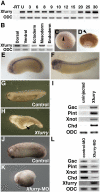
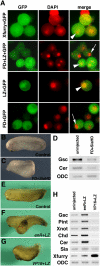
A transcriptional corepressor, Xenopus furry (Xfurry), is expressed in the chordamesodermal region and induces secondary dorsal axes when overexpressed on the ventral side of the embryo. The N-terminal furry domain functions as a repressor, and the C-terminal leucine zipper (LZ) motifs /coiled-coil structure, found only in vertebrate homologs, contributes to the nuclear localization. The engrailed repressor (enR)+LZ repressor construct, which has properties similar to Xfurry, induced several chordamesodermal genes. In contrast, an antisense morpholino oligonucleotide, Xfurry-MO, and the activating construct, herpes simplex virus protein (VP16)+LZ, had effects opposite those of Xfurry overexpression. Because blocking protein synthesis with cycloheximide superinduced several Xfurry transcriptional targets, and because expression of enR+LZ induced such genes under cycloheximide treatment, we analyzed the role of an Xfurry transcriptional target, microRNA miR-15. Cycloheximide reduced the expression of primary miR-15 (pri-miR-15), whereas miR-15 reduced the expression of genes superinduced by cycloheximide treatment. These results show that Xfurry regulates chordamesodermal genes by contributing to repression of pretranscriptional gene silencing by miR-15.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
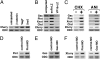

References
-
- Cong J, et al. The furry gene of Drosophila is important for maintaining the integrity of cellular extensions during morphogenesis. Development. 2001;128:2793–2802. - PubMed
-
- Emoto K, et al. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

